Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
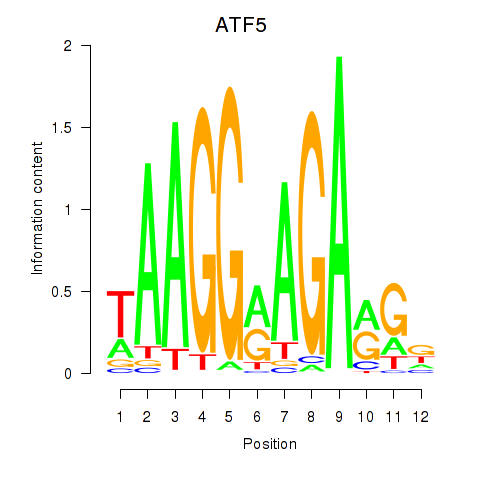
Results for ATF5
Z-value: 0.57
Transcription factors associated with ATF5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF5
|
ENSG00000169136.4 | activating transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF5 | hg19_v2_chr19_+_50431959_50431998 | 0.40 | 4.7e-02 | Click! |
Activity profile of ATF5 motif
Sorted Z-values of ATF5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_4229495 | 3.11 |
ENST00000221847.5
|
EBI3
|
Epstein-Barr virus induced 3 |
| chr3_-_127441406 | 1.87 |
ENST00000487473.1
ENST00000484451.1 |
MGLL
|
monoglyceride lipase |
| chr16_-_65155979 | 1.84 |
ENST00000562325.1
ENST00000268603.4 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr16_-_65155833 | 1.75 |
ENST00000566827.1
ENST00000394156.3 ENST00000562998.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr9_+_116917807 | 1.58 |
ENST00000356083.3
|
COL27A1
|
collagen, type XXVII, alpha 1 |
| chr7_-_115670792 | 1.54 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr7_-_115670804 | 1.47 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr2_-_113542063 | 1.42 |
ENST00000263339.3
|
IL1A
|
interleukin 1, alpha |
| chr7_-_41740181 | 1.41 |
ENST00000442711.1
|
INHBA
|
inhibin, beta A |
| chr4_-_122085469 | 1.09 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr1_+_101361782 | 0.95 |
ENST00000357650.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr9_+_109625378 | 0.90 |
ENST00000277225.5
ENST00000457913.1 ENST00000472574.1 |
ZNF462
|
zinc finger protein 462 |
| chr6_+_43738444 | 0.87 |
ENST00000324450.6
ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA
|
vascular endothelial growth factor A |
| chr2_-_100759037 | 0.83 |
ENST00000317233.4
ENST00000423966.1 ENST00000416492.1 |
AFF3
|
AF4/FMR2 family, member 3 |
| chr10_+_111765562 | 0.78 |
ENST00000360162.3
|
ADD3
|
adducin 3 (gamma) |
| chr19_-_10491234 | 0.75 |
ENST00000524462.1
ENST00000531836.1 ENST00000525621.1 |
TYK2
|
tyrosine kinase 2 |
| chr16_-_72127456 | 0.73 |
ENST00000562153.1
|
TXNL4B
|
thioredoxin-like 4B |
| chr9_+_2622085 | 0.72 |
ENST00000382099.2
|
VLDLR
|
very low density lipoprotein receptor |
| chr9_+_2621798 | 0.71 |
ENST00000382100.3
|
VLDLR
|
very low density lipoprotein receptor |
| chr12_-_16761007 | 0.69 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr5_-_20575959 | 0.68 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr12_+_32655110 | 0.65 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr20_+_58179582 | 0.65 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr14_+_23846328 | 0.64 |
ENST00000382809.2
|
CMTM5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr14_+_32546274 | 0.64 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr6_+_114178512 | 0.64 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr11_-_126870655 | 0.60 |
ENST00000525144.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr1_+_101361626 | 0.56 |
ENST00000370112.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr8_+_22462532 | 0.51 |
ENST00000389279.3
|
CCAR2
|
cell cycle and apoptosis regulator 2 |
| chr2_-_145278475 | 0.49 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr8_+_22462145 | 0.47 |
ENST00000308511.4
ENST00000523801.1 ENST00000521301.1 |
CCAR2
|
cell cycle and apoptosis regulator 2 |
| chr10_-_99531709 | 0.46 |
ENST00000266066.3
|
SFRP5
|
secreted frizzled-related protein 5 |
| chr4_+_77172847 | 0.44 |
ENST00000515604.1
ENST00000539752.1 ENST00000424749.2 |
FAM47E
FAM47E-STBD1
FAM47E
|
uncharacterized protein LOC100631383 FAM47E-STBD1 readthrough family with sequence similarity 47, member E |
| chr1_+_209859510 | 0.43 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr16_-_72128270 | 0.41 |
ENST00000426362.2
|
TXNL4B
|
thioredoxin-like 4B |
| chr2_-_87017985 | 0.41 |
ENST00000352580.3
|
CD8A
|
CD8a molecule |
| chr16_-_30023615 | 0.38 |
ENST00000564979.1
ENST00000563378.1 |
DOC2A
|
double C2-like domains, alpha |
| chr6_+_44095347 | 0.37 |
ENST00000323267.6
|
TMEM63B
|
transmembrane protein 63B |
| chr20_+_62492566 | 0.37 |
ENST00000369916.3
|
ABHD16B
|
abhydrolase domain containing 16B |
| chr11_+_124735282 | 0.35 |
ENST00000397801.1
|
ROBO3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr15_+_63481668 | 0.35 |
ENST00000321437.4
ENST00000559006.1 ENST00000448330.2 |
RAB8B
|
RAB8B, member RAS oncogene family |
| chr7_-_3083573 | 0.34 |
ENST00000396946.4
|
CARD11
|
caspase recruitment domain family, member 11 |
| chr5_+_34757309 | 0.34 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr9_-_38069208 | 0.34 |
ENST00000377707.3
ENST00000377700.4 |
SHB
|
Src homology 2 domain containing adaptor protein B |
| chr17_-_47287928 | 0.33 |
ENST00000507680.1
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr4_+_159443090 | 0.33 |
ENST00000343542.5
ENST00000470033.1 |
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr3_-_8686479 | 0.33 |
ENST00000544814.1
ENST00000427408.1 |
SSUH2
|
ssu-2 homolog (C. elegans) |
| chrX_+_122318113 | 0.33 |
ENST00000371264.3
|
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr14_-_65569186 | 0.32 |
ENST00000555932.1
ENST00000358664.4 ENST00000284165.6 ENST00000358402.4 ENST00000246163.2 ENST00000556979.1 ENST00000555667.1 ENST00000557746.1 ENST00000556443.1 |
MAX
|
MYC associated factor X |
| chrX_+_103810874 | 0.32 |
ENST00000372582.1
|
IL1RAPL2
|
interleukin 1 receptor accessory protein-like 2 |
| chr12_-_49351303 | 0.31 |
ENST00000256682.4
|
ARF3
|
ADP-ribosylation factor 3 |
| chrX_+_153409678 | 0.31 |
ENST00000369951.4
|
OPN1LW
|
opsin 1 (cone pigments), long-wave-sensitive |
| chr1_-_183387723 | 0.30 |
ENST00000287713.6
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr6_-_62996066 | 0.30 |
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr6_-_88411911 | 0.30 |
ENST00000257787.5
|
AKIRIN2
|
akirin 2 |
| chr2_-_87018784 | 0.29 |
ENST00000283635.3
ENST00000538832.1 |
CD8A
|
CD8a molecule |
| chr14_-_65569057 | 0.29 |
ENST00000555419.1
ENST00000341653.2 |
MAX
|
MYC associated factor X |
| chr19_+_15160130 | 0.29 |
ENST00000427043.3
|
CASP14
|
caspase 14, apoptosis-related cysteine peptidase |
| chr16_-_72127550 | 0.28 |
ENST00000268483.3
|
TXNL4B
|
thioredoxin-like 4B |
| chr14_+_23846210 | 0.28 |
ENST00000339180.4
ENST00000342473.4 ENST00000397227.3 ENST00000555731.1 |
CMTM5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr8_-_21645804 | 0.28 |
ENST00000518077.1
ENST00000517892.1 |
GFRA2
|
GDNF family receptor alpha 2 |
| chr17_+_33914276 | 0.27 |
ENST00000592545.1
ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr5_+_50678921 | 0.27 |
ENST00000230658.7
|
ISL1
|
ISL LIM homeobox 1 |
| chr3_+_38206975 | 0.27 |
ENST00000446845.1
ENST00000311806.3 |
OXSR1
|
oxidative stress responsive 1 |
| chr14_-_65569244 | 0.27 |
ENST00000557277.1
ENST00000556892.1 |
MAX
|
MYC associated factor X |
| chr15_-_90222610 | 0.26 |
ENST00000300055.5
|
PLIN1
|
perilipin 1 |
| chr15_-_90222642 | 0.26 |
ENST00000430628.2
|
PLIN1
|
perilipin 1 |
| chr22_-_36236265 | 0.26 |
ENST00000414461.2
ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr5_+_137203465 | 0.26 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chr17_+_33914460 | 0.26 |
ENST00000537622.2
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chrX_+_153448107 | 0.25 |
ENST00000369935.5
|
OPN1MW
|
opsin 1 (cone pigments), medium-wave-sensitive |
| chr18_-_3845293 | 0.25 |
ENST00000400145.2
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr4_+_159443024 | 0.25 |
ENST00000448688.2
|
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr14_+_23845995 | 0.24 |
ENST00000359320.3
|
CMTM5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr20_+_43211149 | 0.24 |
ENST00000372886.1
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr18_-_19748331 | 0.24 |
ENST00000584201.1
|
GATA6-AS1
|
GATA6 antisense RNA 1 (head to head) |
| chr15_-_88799948 | 0.24 |
ENST00000394480.2
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr8_-_95449155 | 0.24 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr22_-_26875345 | 0.24 |
ENST00000398141.1
|
HPS4
|
Hermansky-Pudlak syndrome 4 |
| chr14_-_21566731 | 0.23 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr22_-_26875631 | 0.23 |
ENST00000402105.3
|
HPS4
|
Hermansky-Pudlak syndrome 4 |
| chr5_+_110559603 | 0.23 |
ENST00000512453.1
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr1_-_27930102 | 0.23 |
ENST00000247087.5
ENST00000374011.2 |
AHDC1
|
AT hook, DNA binding motif, containing 1 |
| chr14_-_36988882 | 0.22 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr18_-_3845321 | 0.22 |
ENST00000539435.1
ENST00000400147.2 |
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr3_+_112930387 | 0.21 |
ENST00000485230.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr18_-_19748379 | 0.21 |
ENST00000579431.1
|
GATA6-AS1
|
GATA6 antisense RNA 1 (head to head) |
| chr9_-_23825956 | 0.21 |
ENST00000397312.2
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr5_+_82767487 | 0.21 |
ENST00000343200.5
ENST00000342785.4 |
VCAN
|
versican |
| chr14_+_79745746 | 0.21 |
ENST00000281127.7
|
NRXN3
|
neurexin 3 |
| chr16_+_30075463 | 0.21 |
ENST00000562168.1
ENST00000569545.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr8_-_13134045 | 0.20 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr12_-_100656134 | 0.20 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chrX_+_131157609 | 0.20 |
ENST00000496850.1
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chr12_-_120884175 | 0.20 |
ENST00000546954.1
|
TRIAP1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr15_-_75871589 | 0.19 |
ENST00000306726.2
|
PTPN9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr5_+_156887027 | 0.19 |
ENST00000435489.2
ENST00000311946.7 |
NIPAL4
|
NIPA-like domain containing 4 |
| chr7_-_27142290 | 0.19 |
ENST00000222718.5
|
HOXA2
|
homeobox A2 |
| chr16_-_58663720 | 0.19 |
ENST00000564557.1
ENST00000569240.1 ENST00000441024.2 ENST00000569020.1 ENST00000317147.5 |
CNOT1
|
CCR4-NOT transcription complex, subunit 1 |
| chrX_+_47050236 | 0.18 |
ENST00000377351.4
|
UBA1
|
ubiquitin-like modifier activating enzyme 1 |
| chr17_-_73149921 | 0.17 |
ENST00000481647.1
ENST00000470924.1 |
HN1
|
hematological and neurological expressed 1 |
| chr5_+_82767583 | 0.17 |
ENST00000512590.2
ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN
|
versican |
| chr7_-_155604967 | 0.17 |
ENST00000297261.2
|
SHH
|
sonic hedgehog |
| chr14_+_79746249 | 0.17 |
ENST00000428277.2
|
NRXN3
|
neurexin 3 |
| chr3_-_182698381 | 0.17 |
ENST00000292782.4
|
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chrX_+_133930798 | 0.17 |
ENST00000414371.2
|
FAM122C
|
family with sequence similarity 122C |
| chr1_-_151804222 | 0.17 |
ENST00000392697.3
|
RORC
|
RAR-related orphan receptor C |
| chr4_-_159956333 | 0.17 |
ENST00000434826.2
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr4_-_175750364 | 0.16 |
ENST00000340217.5
ENST00000274093.3 |
GLRA3
|
glycine receptor, alpha 3 |
| chr2_-_214016314 | 0.16 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr8_+_79428539 | 0.16 |
ENST00000352966.5
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chrX_-_47489244 | 0.16 |
ENST00000469388.1
ENST00000396992.3 ENST00000377005.2 |
CFP
|
complement factor properdin |
| chr6_+_87865262 | 0.16 |
ENST00000369577.3
ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr8_-_61193947 | 0.16 |
ENST00000317995.4
|
CA8
|
carbonic anhydrase VIII |
| chr9_+_103947311 | 0.15 |
ENST00000395056.2
|
LPPR1
|
Lipid phosphate phosphatase-related protein type 1 |
| chr14_+_32546145 | 0.15 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr11_+_62495997 | 0.15 |
ENST00000316461.4
|
TTC9C
|
tetratricopeptide repeat domain 9C |
| chr5_+_154393260 | 0.14 |
ENST00000435029.4
|
KIF4B
|
kinesin family member 4B |
| chr11_-_132813566 | 0.14 |
ENST00000331898.7
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr8_-_623547 | 0.14 |
ENST00000522893.1
|
ERICH1
|
glutamate-rich 1 |
| chr5_-_134788086 | 0.14 |
ENST00000537858.1
|
TIFAB
|
TRAF-interacting protein with forkhead-associated domain, family member B |
| chr6_+_72596604 | 0.14 |
ENST00000348717.5
ENST00000517960.1 ENST00000518273.1 ENST00000522291.1 ENST00000521978.1 ENST00000520567.1 ENST00000264839.7 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr13_+_28366780 | 0.14 |
ENST00000302945.2
|
GSX1
|
GS homeobox 1 |
| chr12_+_120884222 | 0.13 |
ENST00000551765.1
ENST00000229384.5 |
GATC
|
glutamyl-tRNA(Gln) amidotransferase, subunit C |
| chr19_-_7936344 | 0.13 |
ENST00000599142.1
|
CTD-3193O13.9
|
Protein FLJ22184 |
| chr11_-_108408895 | 0.13 |
ENST00000443411.1
ENST00000533052.1 |
EXPH5
|
exophilin 5 |
| chr1_-_27286897 | 0.13 |
ENST00000320567.5
|
C1orf172
|
chromosome 1 open reading frame 172 |
| chr1_+_48688357 | 0.13 |
ENST00000533824.1
ENST00000438567.2 ENST00000236495.5 ENST00000420136.2 |
SLC5A9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr15_+_80733570 | 0.13 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr1_+_206579736 | 0.13 |
ENST00000439126.1
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr16_+_22103828 | 0.13 |
ENST00000567131.1
ENST00000568328.1 ENST00000389398.5 ENST00000389397.4 |
VWA3A
|
von Willebrand factor A domain containing 3A |
| chr11_-_106889250 | 0.12 |
ENST00000526355.2
|
GUCY1A2
|
guanylate cyclase 1, soluble, alpha 2 |
| chrX_+_122318006 | 0.12 |
ENST00000371266.1
ENST00000264357.5 |
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr14_-_23877474 | 0.12 |
ENST00000405093.3
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr17_-_6947225 | 0.12 |
ENST00000574600.1
ENST00000308009.1 ENST00000447225.1 |
SLC16A11
|
solute carrier family 16, member 11 |
| chr5_+_132009675 | 0.12 |
ENST00000231449.2
ENST00000350025.2 |
IL4
|
interleukin 4 |
| chr7_-_102232891 | 0.12 |
ENST00000514917.2
|
RP11-514P8.7
|
RP11-514P8.7 |
| chr11_+_62496114 | 0.11 |
ENST00000532583.1
|
TTC9C
|
tetratricopeptide repeat domain 9C |
| chr11_-_106889157 | 0.11 |
ENST00000282249.2
|
GUCY1A2
|
guanylate cyclase 1, soluble, alpha 2 |
| chr16_+_28943260 | 0.11 |
ENST00000538922.1
ENST00000324662.3 ENST00000567541.1 |
CD19
|
CD19 molecule |
| chr16_+_57438679 | 0.11 |
ENST00000219244.4
|
CCL17
|
chemokine (C-C motif) ligand 17 |
| chr12_-_72057638 | 0.11 |
ENST00000552037.1
ENST00000378743.3 |
ZFC3H1
|
zinc finger, C3H1-type containing |
| chr19_-_8642289 | 0.11 |
ENST00000596675.1
ENST00000338257.8 |
MYO1F
|
myosin IF |
| chr14_-_21567009 | 0.10 |
ENST00000556174.1
ENST00000554478.1 ENST00000553980.1 ENST00000421093.2 |
ZNF219
|
zinc finger protein 219 |
| chr5_-_132200477 | 0.10 |
ENST00000296875.2
|
GDF9
|
growth differentiation factor 9 |
| chr5_+_82767284 | 0.10 |
ENST00000265077.3
|
VCAN
|
versican |
| chr19_+_10531150 | 0.10 |
ENST00000352831.6
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr2_-_114514181 | 0.10 |
ENST00000409342.1
|
SLC35F5
|
solute carrier family 35, member F5 |
| chr5_+_148786518 | 0.10 |
ENST00000518014.1
ENST00000505340.1 ENST00000509909.1 |
AC131025.8
MIR143HG
|
AC131025.8 MIR143 host gene (non-protein coding) |
| chr4_+_84377198 | 0.10 |
ENST00000507349.1
ENST00000509970.1 ENST00000505719.1 |
MRPS18C
|
mitochondrial ribosomal protein S18C |
| chr12_-_57030115 | 0.09 |
ENST00000379441.3
ENST00000179765.5 ENST00000551812.1 |
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr1_-_54872059 | 0.09 |
ENST00000371320.3
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr19_-_18366182 | 0.09 |
ENST00000355502.3
|
PDE4C
|
phosphodiesterase 4C, cAMP-specific |
| chr22_-_36357671 | 0.08 |
ENST00000408983.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr3_-_47950745 | 0.08 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr18_-_24128496 | 0.08 |
ENST00000417602.1
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr3_-_88108212 | 0.08 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr1_+_3607228 | 0.08 |
ENST00000378285.1
ENST00000378280.1 ENST00000378288.4 |
TP73
|
tumor protein p73 |
| chr10_-_102989551 | 0.07 |
ENST00000370193.2
|
LBX1
|
ladybird homeobox 1 |
| chrX_-_13835147 | 0.07 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr3_+_101504200 | 0.07 |
ENST00000422132.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr9_-_23826298 | 0.07 |
ENST00000380117.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr8_+_37553261 | 0.07 |
ENST00000331569.4
|
ZNF703
|
zinc finger protein 703 |
| chr6_-_44095183 | 0.06 |
ENST00000372014.3
|
MRPL14
|
mitochondrial ribosomal protein L14 |
| chr6_-_132967142 | 0.06 |
ENST00000275216.1
|
TAAR1
|
trace amine associated receptor 1 |
| chr6_+_30297306 | 0.06 |
ENST00000420746.1
ENST00000513556.1 |
TRIM39
TRIM39-RPP21
|
tripartite motif containing 39 TRIM39-RPP21 readthrough |
| chr17_+_47287749 | 0.06 |
ENST00000419580.2
|
ABI3
|
ABI family, member 3 |
| chr17_+_4675175 | 0.06 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr12_+_104680659 | 0.06 |
ENST00000526691.1
ENST00000531691.1 ENST00000388854.3 ENST00000354940.6 ENST00000526390.1 ENST00000531689.1 |
TXNRD1
|
thioredoxin reductase 1 |
| chr12_+_10124001 | 0.06 |
ENST00000396507.3
ENST00000304361.4 ENST00000434319.2 |
CLEC12A
|
C-type lectin domain family 12, member A |
| chr1_+_202091980 | 0.06 |
ENST00000367282.5
|
GPR37L1
|
G protein-coupled receptor 37 like 1 |
| chrX_+_122318224 | 0.05 |
ENST00000542149.1
|
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr3_+_112930306 | 0.05 |
ENST00000495514.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr5_-_39219705 | 0.05 |
ENST00000351578.6
|
FYB
|
FYN binding protein |
| chr2_-_26700900 | 0.05 |
ENST00000338581.6
ENST00000339598.3 ENST00000402415.3 |
OTOF
|
otoferlin |
| chr10_-_75415825 | 0.05 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr6_+_29079668 | 0.04 |
ENST00000377169.1
|
OR2J3
|
olfactory receptor, family 2, subfamily J, member 3 |
| chr8_-_131028869 | 0.04 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr14_+_50065376 | 0.04 |
ENST00000298288.6
|
LRR1
|
leucine rich repeat protein 1 |
| chr13_-_36944307 | 0.04 |
ENST00000355182.4
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr2_-_128642434 | 0.04 |
ENST00000393001.1
|
AMMECR1L
|
AMMECR1-like |
| chr8_+_145294015 | 0.04 |
ENST00000544576.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr12_-_115121962 | 0.04 |
ENST00000349155.2
|
TBX3
|
T-box 3 |
| chrX_+_122318318 | 0.04 |
ENST00000371251.1
ENST00000371256.5 |
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr2_-_128784846 | 0.04 |
ENST00000259235.3
ENST00000357702.5 ENST00000424298.1 |
SAP130
|
Sin3A-associated protein, 130kDa |
| chr22_-_36236623 | 0.04 |
ENST00000405409.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr1_-_156460391 | 0.04 |
ENST00000360595.3
|
MEF2D
|
myocyte enhancer factor 2D |
| chr1_-_182641037 | 0.03 |
ENST00000483095.2
|
RGS8
|
regulator of G-protein signaling 8 |
| chr4_+_159442878 | 0.03 |
ENST00000307765.5
ENST00000423548.1 |
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr2_-_190927447 | 0.03 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr12_-_72057571 | 0.03 |
ENST00000548100.1
|
ZFC3H1
|
zinc finger, C3H1-type containing |
| chr1_+_158323755 | 0.03 |
ENST00000368157.1
ENST00000368156.1 ENST00000368155.3 ENST00000368154.1 ENST00000368160.3 ENST00000368161.3 |
CD1E
|
CD1e molecule |
| chr12_-_54778471 | 0.03 |
ENST00000550120.1
ENST00000394313.2 ENST00000547210.1 |
ZNF385A
|
zinc finger protein 385A |
| chr1_+_36023035 | 0.03 |
ENST00000373253.3
|
NCDN
|
neurochondrin |
| chr17_+_55055466 | 0.03 |
ENST00000262288.3
ENST00000572710.1 ENST00000575395.1 |
SCPEP1
|
serine carboxypeptidase 1 |
| chr1_-_182640988 | 0.03 |
ENST00000367556.1
|
RGS8
|
regulator of G-protein signaling 8 |
| chr17_+_41150290 | 0.03 |
ENST00000589037.1
ENST00000253788.5 |
RPL27
|
ribosomal protein L27 |
| chr5_-_156390230 | 0.02 |
ENST00000407087.3
ENST00000274532.2 |
TIMD4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chr19_-_45873642 | 0.02 |
ENST00000485403.2
ENST00000586856.1 ENST00000586131.1 ENST00000391940.4 ENST00000221481.6 ENST00000391944.3 ENST00000391945.4 |
ERCC2
|
excision repair cross-complementing rodent repair deficiency, complementation group 2 |
| chr10_-_94003003 | 0.02 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.3 | 4.1 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.3 | 0.9 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.3 | 1.9 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 3.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.2 | 1.4 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.2 | 1.4 | GO:2000391 | positive regulation of neutrophil extravasation(GO:2000391) |
| 0.2 | 1.6 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.2 | 0.7 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.2 | 0.5 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.2 | 1.5 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.5 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.1 | 0.7 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.5 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 0.3 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) cardiac cell fate determination(GO:0060913) |
| 0.1 | 0.3 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.3 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.2 | GO:0019056 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.1 | 0.6 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.1 | 1.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.2 | GO:0035283 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.2 | GO:0060738 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of immature T cell proliferation in thymus(GO:0033092) right lung development(GO:0060458) primary prostatic bud elongation(GO:0060516) epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.1 | 0.3 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.3 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.1 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.4 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.1 | GO:0071288 | T-helper 1 cell lineage commitment(GO:0002296) B cell costimulation(GO:0031296) cellular response to mercury ion(GO:0071288) negative regulation of complement-dependent cytotoxicity(GO:1903660) positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.3 | GO:0070970 | interleukin-2 secretion(GO:0070970) |
| 0.0 | 0.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.0 | 0.1 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.4 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 1.0 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 | 0.4 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.2 | GO:2001138 | regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.0 | 0.5 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 1.4 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 3.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.0 | GO:0060931 | sinoatrial node development(GO:0003163) sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.0 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.0 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.9 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.2 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.0 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.3 | 1.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 0.6 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.5 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 1.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.2 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 1.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 1.4 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.3 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 1.0 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.5 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.9 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.6 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.5 | 1.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.9 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.4 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 1.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.3 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 1.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.3 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 1.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.4 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.2 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.3 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.7 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 3.1 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.1 | GO:0098625 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.0 | 1.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.9 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.0 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |