Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
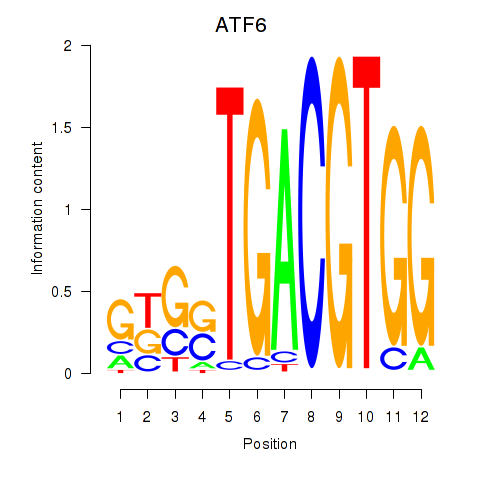
Results for ATF6
Z-value: 0.58
Transcription factors associated with ATF6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF6
|
ENSG00000118217.5 | activating transcription factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF6 | hg19_v2_chr1_+_161736072_161736093 | 0.06 | 7.8e-01 | Click! |
Activity profile of ATF6 motif
Sorted Z-values of ATF6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_131762105 | 1.38 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr18_+_77623668 | 0.89 |
ENST00000316249.3
|
KCNG2
|
potassium voltage-gated channel, subfamily G, member 2 |
| chr13_-_49107303 | 0.87 |
ENST00000344532.3
|
RCBTB2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr16_+_53164833 | 0.83 |
ENST00000564845.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr17_-_49337392 | 0.67 |
ENST00000376381.2
ENST00000586178.1 |
MBTD1
|
mbt domain containing 1 |
| chr9_+_71320557 | 0.62 |
ENST00000541509.1
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| chr11_-_119234876 | 0.60 |
ENST00000525735.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr1_+_221051699 | 0.60 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr8_-_8243968 | 0.57 |
ENST00000520004.1
|
SGK223
|
Tyrosine-protein kinase SgK223 |
| chr21_+_38445539 | 0.52 |
ENST00000418766.1
ENST00000450533.1 ENST00000438055.1 ENST00000355666.1 ENST00000540756.1 ENST00000399010.1 |
TTC3
|
tetratricopeptide repeat domain 3 |
| chr12_+_56661033 | 0.52 |
ENST00000433805.2
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr12_+_56660633 | 0.51 |
ENST00000308197.5
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr2_+_219745020 | 0.51 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr16_+_69373471 | 0.49 |
ENST00000569637.2
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr4_-_39640513 | 0.49 |
ENST00000511809.1
ENST00000505729.1 |
SMIM14
|
small integral membrane protein 14 |
| chr19_-_42759300 | 0.48 |
ENST00000222329.4
|
ERF
|
Ets2 repressor factor |
| chr17_-_66951474 | 0.47 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr6_+_24775641 | 0.42 |
ENST00000378054.2
ENST00000476555.1 |
GMNN
|
geminin, DNA replication inhibitor |
| chr3_+_184529948 | 0.41 |
ENST00000436792.2
ENST00000446204.2 ENST00000422105.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr3_+_184529929 | 0.41 |
ENST00000287546.4
ENST00000437079.3 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr14_-_23822061 | 0.41 |
ENST00000397260.3
|
SLC22A17
|
solute carrier family 22, member 17 |
| chr14_-_23822080 | 0.40 |
ENST00000397267.1
ENST00000354772.3 |
SLC22A17
|
solute carrier family 22, member 17 |
| chr6_+_132891461 | 0.40 |
ENST00000275198.1
|
TAAR6
|
trace amine associated receptor 6 |
| chr8_+_29952914 | 0.40 |
ENST00000321250.8
ENST00000518001.1 ENST00000520682.1 ENST00000442880.2 ENST00000523116.1 |
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chr1_-_211752073 | 0.39 |
ENST00000367001.4
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1 |
| chr19_+_38810447 | 0.39 |
ENST00000263372.3
|
KCNK6
|
potassium channel, subfamily K, member 6 |
| chr1_+_222791417 | 0.38 |
ENST00000344922.5
ENST00000344441.6 ENST00000344507.1 |
MIA3
|
melanoma inhibitory activity family, member 3 |
| chr3_-_33759699 | 0.38 |
ENST00000399362.4
ENST00000359576.5 ENST00000307312.7 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr3_-_33759541 | 0.38 |
ENST00000468888.2
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr5_+_41904431 | 0.38 |
ENST00000381647.2
|
C5orf51
|
chromosome 5 open reading frame 51 |
| chr7_+_16793160 | 0.38 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr5_+_78532003 | 0.38 |
ENST00000396137.4
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr17_+_6554971 | 0.37 |
ENST00000391428.2
|
C17orf100
|
chromosome 17 open reading frame 100 |
| chr21_-_38445470 | 0.36 |
ENST00000399098.1
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr15_-_23692381 | 0.36 |
ENST00000567107.1
ENST00000345070.5 ENST00000312015.5 |
GOLGA6L2
|
golgin A6 family-like 2 |
| chr1_-_15850676 | 0.36 |
ENST00000440484.1
ENST00000333868.5 |
CASP9
|
caspase 9, apoptosis-related cysteine peptidase |
| chr17_+_66511540 | 0.36 |
ENST00000588188.2
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr21_-_38445297 | 0.35 |
ENST00000430792.1
ENST00000399103.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr2_-_105946491 | 0.34 |
ENST00000393359.2
|
TGFBRAP1
|
transforming growth factor, beta receptor associated protein 1 |
| chr21_-_38445011 | 0.34 |
ENST00000464265.1
ENST00000399102.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr17_+_65821780 | 0.34 |
ENST00000321892.4
ENST00000335221.5 ENST00000306378.6 |
BPTF
|
bromodomain PHD finger transcription factor |
| chr14_+_68086515 | 0.33 |
ENST00000261783.3
|
ARG2
|
arginase 2 |
| chr7_-_100808843 | 0.33 |
ENST00000249330.2
|
VGF
|
VGF nerve growth factor inducible |
| chr6_+_127588020 | 0.33 |
ENST00000309649.3
ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146
|
ring finger protein 146 |
| chr7_-_86849883 | 0.33 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr11_-_77791156 | 0.32 |
ENST00000281031.4
|
NDUFC2
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa |
| chr1_-_78149041 | 0.32 |
ENST00000414381.1
ENST00000370798.1 |
ZZZ3
|
zinc finger, ZZ-type containing 3 |
| chr4_-_119757239 | 0.32 |
ENST00000280551.6
|
SEC24D
|
SEC24 family member D |
| chr13_-_101327028 | 0.32 |
ENST00000328767.5
ENST00000342624.5 ENST00000376234.3 ENST00000423847.1 |
TMTC4
|
transmembrane and tetratricopeptide repeat containing 4 |
| chr5_+_68389807 | 0.31 |
ENST00000380860.4
ENST00000504103.1 ENST00000502979.1 |
SLC30A5
|
solute carrier family 30 (zinc transporter), member 5 |
| chr10_+_102759045 | 0.31 |
ENST00000370220.1
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr10_-_94333784 | 0.31 |
ENST00000265986.6
|
IDE
|
insulin-degrading enzyme |
| chr8_-_38126635 | 0.31 |
ENST00000529359.1
|
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr1_-_25573977 | 0.31 |
ENST00000243189.7
|
C1orf63
|
chromosome 1 open reading frame 63 |
| chr1_-_25573937 | 0.31 |
ENST00000417642.2
ENST00000431849.2 |
C1orf63
|
chromosome 1 open reading frame 63 |
| chr6_-_6007200 | 0.31 |
ENST00000244766.2
|
NRN1
|
neuritin 1 |
| chr17_-_39968406 | 0.31 |
ENST00000393928.1
|
LEPREL4
|
leprecan-like 4 |
| chr15_+_96869165 | 0.31 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr4_-_24914576 | 0.30 |
ENST00000502801.1
ENST00000428116.2 |
CCDC149
|
coiled-coil domain containing 149 |
| chr8_+_29953163 | 0.30 |
ENST00000518192.1
|
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chr22_-_39548627 | 0.30 |
ENST00000216133.5
|
CBX7
|
chromobox homolog 7 |
| chr7_-_100808394 | 0.29 |
ENST00000445482.2
|
VGF
|
VGF nerve growth factor inducible |
| chr1_+_156830607 | 0.29 |
ENST00000368196.3
|
NTRK1
|
neurotrophic tyrosine kinase, receptor, type 1 |
| chr7_+_44646162 | 0.29 |
ENST00000439616.2
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr12_+_147052 | 0.29 |
ENST00000594563.1
|
AC026369.1
|
Uncharacterized protein |
| chr20_-_34638841 | 0.29 |
ENST00000565493.1
|
LINC00657
|
long intergenic non-protein coding RNA 657 |
| chr6_+_127587755 | 0.28 |
ENST00000368314.1
ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146
|
ring finger protein 146 |
| chr1_+_156830678 | 0.28 |
ENST00000524377.1
ENST00000358660.3 |
NTRK1
|
neurotrophic tyrosine kinase, receptor, type 1 |
| chr11_+_28131821 | 0.27 |
ENST00000379199.2
ENST00000303459.6 |
METTL15
|
methyltransferase like 15 |
| chr7_+_116593568 | 0.27 |
ENST00000446490.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr3_+_100211412 | 0.27 |
ENST00000323523.4
ENST00000403410.1 ENST00000449609.1 |
TMEM45A
|
transmembrane protein 45A |
| chr17_+_65821636 | 0.27 |
ENST00000544778.2
|
BPTF
|
bromodomain PHD finger transcription factor |
| chr14_-_64970494 | 0.27 |
ENST00000608382.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr12_-_110011288 | 0.26 |
ENST00000540016.1
ENST00000266839.5 |
MMAB
|
methylmalonic aciduria (cobalamin deficiency) cblB type |
| chr13_-_36920615 | 0.25 |
ENST00000494062.2
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr7_+_116593433 | 0.25 |
ENST00000323984.3
ENST00000393449.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr10_+_35415719 | 0.25 |
ENST00000474362.1
ENST00000374721.3 |
CREM
|
cAMP responsive element modulator |
| chr7_+_12726474 | 0.25 |
ENST00000396662.1
ENST00000356797.3 ENST00000396664.2 |
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr1_-_15850839 | 0.25 |
ENST00000348549.5
ENST00000546424.1 |
CASP9
|
caspase 9, apoptosis-related cysteine peptidase |
| chr11_+_28129795 | 0.24 |
ENST00000406787.3
ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15
|
methyltransferase like 15 |
| chr21_-_38445443 | 0.24 |
ENST00000360525.4
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr1_+_33005020 | 0.24 |
ENST00000373510.4
ENST00000316459.4 |
ZBTB8A
|
zinc finger and BTB domain containing 8A |
| chr9_+_133978190 | 0.24 |
ENST00000372312.3
|
AIF1L
|
allograft inflammatory factor 1-like |
| chr1_-_161993616 | 0.24 |
ENST00000294794.3
|
OLFML2B
|
olfactomedin-like 2B |
| chr10_+_112257596 | 0.24 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5 |
| chr4_-_39640700 | 0.23 |
ENST00000295958.5
|
SMIM14
|
small integral membrane protein 14 |
| chr9_-_127703333 | 0.23 |
ENST00000373555.4
|
GOLGA1
|
golgin A1 |
| chr14_+_105190514 | 0.23 |
ENST00000330877.2
|
ADSSL1
|
adenylosuccinate synthase like 1 |
| chr11_-_77790865 | 0.23 |
ENST00000534029.1
ENST00000525085.1 ENST00000527806.1 ENST00000528164.1 ENST00000528251.1 ENST00000530054.1 |
NDUFC2
NDUFC2-KCTD14
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa NDUFC2-KCTD14 readthrough |
| chr17_-_57184260 | 0.23 |
ENST00000376149.3
ENST00000393066.3 |
TRIM37
|
tripartite motif containing 37 |
| chr1_+_185126291 | 0.23 |
ENST00000367500.4
|
SWT1
|
SWT1 RNA endoribonuclease homolog (S. cerevisiae) |
| chr15_-_52030293 | 0.23 |
ENST00000560491.1
ENST00000267838.3 |
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr5_+_99871004 | 0.22 |
ENST00000312637.4
|
FAM174A
|
family with sequence similarity 174, member A |
| chr9_-_77643307 | 0.22 |
ENST00000376834.3
ENST00000376830.3 |
C9orf41
|
chromosome 9 open reading frame 41 |
| chr2_-_211036051 | 0.22 |
ENST00000418791.1
ENST00000452086.1 ENST00000281772.9 |
KANSL1L
|
KAT8 regulatory NSL complex subunit 1-like |
| chr9_+_71320596 | 0.22 |
ENST00000265382.3
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| chr22_+_38864041 | 0.22 |
ENST00000216014.4
ENST00000409006.3 |
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr4_+_128982430 | 0.22 |
ENST00000512292.1
ENST00000508819.1 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr17_-_39968855 | 0.22 |
ENST00000355468.3
ENST00000590496.1 |
LEPREL4
|
leprecan-like 4 |
| chr8_+_97657449 | 0.22 |
ENST00000220763.5
|
CPQ
|
carboxypeptidase Q |
| chr11_+_20409070 | 0.21 |
ENST00000331079.6
|
PRMT3
|
protein arginine methyltransferase 3 |
| chr4_+_128982416 | 0.21 |
ENST00000326639.6
|
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr4_-_169931393 | 0.21 |
ENST00000504480.1
ENST00000306193.3 |
CBR4
|
carbonyl reductase 4 |
| chr2_+_28615669 | 0.21 |
ENST00000379619.1
ENST00000264716.4 |
FOSL2
|
FOS-like antigen 2 |
| chr9_-_77643189 | 0.20 |
ENST00000376837.3
|
C9orf41
|
chromosome 9 open reading frame 41 |
| chr1_-_113498616 | 0.20 |
ENST00000433570.4
ENST00000538576.1 ENST00000458229.1 |
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr10_+_35416090 | 0.20 |
ENST00000354759.3
|
CREM
|
cAMP responsive element modulator |
| chr1_-_78148324 | 0.20 |
ENST00000370801.3
ENST00000433749.1 |
ZZZ3
|
zinc finger, ZZ-type containing 3 |
| chr9_+_133454943 | 0.20 |
ENST00000319725.9
|
FUBP3
|
far upstream element (FUSE) binding protein 3 |
| chr5_+_122110691 | 0.20 |
ENST00000379516.2
ENST00000505934.1 ENST00000514949.1 |
SNX2
|
sorting nexin 2 |
| chr10_+_23728198 | 0.20 |
ENST00000376495.3
|
OTUD1
|
OTU domain containing 1 |
| chr11_-_118927816 | 0.20 |
ENST00000534233.1
ENST00000532752.1 ENST00000525859.1 ENST00000404233.3 ENST00000532421.1 ENST00000543287.1 ENST00000527310.2 ENST00000529972.1 |
HYOU1
|
hypoxia up-regulated 1 |
| chr10_+_35415978 | 0.19 |
ENST00000429130.3
ENST00000469949.2 ENST00000460270.1 |
CREM
|
cAMP responsive element modulator |
| chr6_-_84937314 | 0.19 |
ENST00000257766.4
ENST00000403245.3 |
KIAA1009
|
KIAA1009 |
| chr9_-_79520989 | 0.19 |
ENST00000376713.3
ENST00000376718.3 ENST00000428286.1 |
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr19_+_2269485 | 0.19 |
ENST00000582888.4
ENST00000602676.2 ENST00000322297.4 ENST00000583542.4 |
OAZ1
|
ornithine decarboxylase antizyme 1 |
| chr14_-_35183755 | 0.19 |
ENST00000555765.1
|
CFL2
|
cofilin 2 (muscle) |
| chr4_-_99850243 | 0.19 |
ENST00000280892.6
ENST00000511644.1 ENST00000504432.1 ENST00000505992.1 |
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr9_+_36036430 | 0.19 |
ENST00000377966.3
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr13_+_114238997 | 0.19 |
ENST00000538138.1
ENST00000375370.5 |
TFDP1
|
transcription factor Dp-1 |
| chr4_-_24914508 | 0.19 |
ENST00000504487.1
|
CCDC149
|
coiled-coil domain containing 149 |
| chr17_-_15466850 | 0.19 |
ENST00000438826.3
ENST00000225576.3 ENST00000519970.1 ENST00000518321.1 ENST00000428082.2 ENST00000522212.2 |
TVP23C
TVP23C-CDRT4
|
trans-golgi network vesicle protein 23 homolog C (S. cerevisiae) TVP23C-CDRT4 readthrough |
| chr18_+_56530794 | 0.18 |
ENST00000590285.1
ENST00000586085.1 ENST00000589288.1 |
ZNF532
|
zinc finger protein 532 |
| chr8_-_38126675 | 0.18 |
ENST00000531823.1
ENST00000534339.1 ENST00000524616.1 ENST00000422581.2 ENST00000424479.2 ENST00000419686.2 |
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr5_+_94890778 | 0.18 |
ENST00000380009.4
|
ARSK
|
arylsulfatase family, member K |
| chr9_-_131709858 | 0.17 |
ENST00000372586.3
|
DOLK
|
dolichol kinase |
| chr8_-_75233563 | 0.17 |
ENST00000342232.4
|
JPH1
|
junctophilin 1 |
| chr7_+_44646177 | 0.17 |
ENST00000443864.2
ENST00000447398.1 ENST00000449767.1 ENST00000419661.1 |
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr12_+_57853918 | 0.17 |
ENST00000532291.1
ENST00000543426.1 ENST00000228682.2 ENST00000546141.1 |
GLI1
|
GLI family zinc finger 1 |
| chr19_-_42927251 | 0.17 |
ENST00000597001.1
|
LIPE
|
lipase, hormone-sensitive |
| chr20_+_39657454 | 0.17 |
ENST00000361337.2
|
TOP1
|
topoisomerase (DNA) I |
| chr17_+_7211656 | 0.17 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr6_+_24775153 | 0.17 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr13_-_36920872 | 0.16 |
ENST00000451493.1
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr11_-_35547151 | 0.16 |
ENST00000378878.3
ENST00000529303.1 ENST00000278360.3 |
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr4_+_75310851 | 0.16 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr14_+_50087468 | 0.16 |
ENST00000305386.2
|
MGAT2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr7_+_128379449 | 0.16 |
ENST00000479257.1
|
CALU
|
calumenin |
| chr12_+_1100370 | 0.16 |
ENST00000543086.3
ENST00000546231.2 ENST00000397203.2 |
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr14_+_103388976 | 0.16 |
ENST00000299155.5
|
AMN
|
amnion associated transmembrane protein |
| chr9_+_131038425 | 0.15 |
ENST00000320188.5
ENST00000608796.1 ENST00000419867.2 ENST00000418976.1 |
SWI5
|
SWI5 recombination repair homolog (yeast) |
| chr13_-_36920420 | 0.15 |
ENST00000438666.2
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr9_+_71394945 | 0.15 |
ENST00000394264.3
|
FAM122A
|
family with sequence similarity 122A |
| chr20_+_34042962 | 0.15 |
ENST00000446710.1
ENST00000420564.1 |
CEP250
|
centrosomal protein 250kDa |
| chr6_-_151773232 | 0.15 |
ENST00000444024.1
ENST00000367303.4 |
RMND1
|
required for meiotic nuclear division 1 homolog (S. cerevisiae) |
| chrX_-_100872911 | 0.15 |
ENST00000361910.4
ENST00000539247.1 ENST00000538627.1 |
ARMCX6
|
armadillo repeat containing, X-linked 6 |
| chr6_+_159290917 | 0.15 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr2_+_183580954 | 0.15 |
ENST00000264065.7
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr7_+_44646218 | 0.15 |
ENST00000444676.1
ENST00000222673.5 |
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr14_-_77495007 | 0.15 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr4_-_1242764 | 0.15 |
ENST00000514210.1
ENST00000382952.3 ENST00000290921.6 |
CTBP1
|
C-terminal binding protein 1 |
| chr14_-_35183886 | 0.15 |
ENST00000298159.6
|
CFL2
|
cofilin 2 (muscle) |
| chr1_-_185126037 | 0.15 |
ENST00000367506.5
ENST00000367504.3 |
TRMT1L
|
tRNA methyltransferase 1 homolog (S. cerevisiae)-like |
| chr6_-_33756867 | 0.14 |
ENST00000293760.5
|
LEMD2
|
LEM domain containing 2 |
| chr5_-_171881491 | 0.14 |
ENST00000311601.5
|
SH3PXD2B
|
SH3 and PX domains 2B |
| chr12_-_76953453 | 0.14 |
ENST00000549570.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr2_-_40006289 | 0.14 |
ENST00000260619.6
ENST00000454352.2 |
THUMPD2
|
THUMP domain containing 2 |
| chrX_-_128977781 | 0.14 |
ENST00000357166.6
|
ZDHHC9
|
zinc finger, DHHC-type containing 9 |
| chr1_+_87794150 | 0.14 |
ENST00000370544.5
|
LMO4
|
LIM domain only 4 |
| chr17_-_19771242 | 0.14 |
ENST00000361658.2
|
ULK2
|
unc-51 like autophagy activating kinase 2 |
| chr14_+_64970662 | 0.13 |
ENST00000556965.1
ENST00000554015.1 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr11_-_28129656 | 0.13 |
ENST00000263181.6
|
KIF18A
|
kinesin family member 18A |
| chr4_+_108745711 | 0.13 |
ENST00000394684.4
|
SGMS2
|
sphingomyelin synthase 2 |
| chr2_+_242254507 | 0.13 |
ENST00000391973.2
|
SEPT2
|
septin 2 |
| chr4_+_141264597 | 0.13 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chrX_-_100914781 | 0.13 |
ENST00000431597.1
ENST00000458024.1 ENST00000413506.1 ENST00000440675.1 ENST00000328766.5 ENST00000356824.4 |
ARMCX2
|
armadillo repeat containing, X-linked 2 |
| chr15_+_76135622 | 0.13 |
ENST00000338677.4
ENST00000267938.4 ENST00000569423.1 |
UBE2Q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr3_+_183415558 | 0.13 |
ENST00000305135.5
|
YEATS2
|
YEATS domain containing 2 |
| chr16_-_86588627 | 0.13 |
ENST00000565482.1
ENST00000564364.1 ENST00000561989.1 ENST00000543303.2 ENST00000381214.5 ENST00000360900.6 ENST00000322911.6 ENST00000546093.1 ENST00000569000.1 ENST00000562994.1 ENST00000561522.1 |
MTHFSD
|
methenyltetrahydrofolate synthetase domain containing |
| chr1_-_193028632 | 0.13 |
ENST00000421683.1
|
UCHL5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr3_-_121468513 | 0.13 |
ENST00000494517.1
ENST00000393667.3 |
GOLGB1
|
golgin B1 |
| chr10_+_121410882 | 0.13 |
ENST00000369085.3
|
BAG3
|
BCL2-associated athanogene 3 |
| chr1_+_87170577 | 0.13 |
ENST00000482504.1
|
SH3GLB1
|
SH3-domain GRB2-like endophilin B1 |
| chr4_+_668348 | 0.13 |
ENST00000511290.1
|
MYL5
|
myosin, light chain 5, regulatory |
| chr6_+_159291090 | 0.12 |
ENST00000367073.4
ENST00000608817.1 |
C6orf99
|
chromosome 6 open reading frame 99 |
| chr10_-_104953009 | 0.12 |
ENST00000470299.1
ENST00000343289.5 |
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr7_+_92861653 | 0.12 |
ENST00000251739.5
ENST00000305866.5 ENST00000544910.1 ENST00000541136.1 ENST00000458530.1 ENST00000535481.1 ENST00000317751.6 |
CCDC132
|
coiled-coil domain containing 132 |
| chr9_+_35538616 | 0.12 |
ENST00000455600.1
|
RUSC2
|
RUN and SH3 domain containing 2 |
| chr2_-_190649082 | 0.12 |
ENST00000392350.3
ENST00000392349.4 |
ORMDL1
|
ORM1-like 1 (S. cerevisiae) |
| chr1_-_193028426 | 0.12 |
ENST00000367450.3
ENST00000530098.2 ENST00000367451.4 ENST00000367448.1 ENST00000367449.1 |
UCHL5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr3_-_121468602 | 0.12 |
ENST00000340645.5
|
GOLGB1
|
golgin B1 |
| chr2_+_242254679 | 0.12 |
ENST00000428282.1
ENST00000360051.3 |
SEPT2
|
septin 2 |
| chr1_+_202976493 | 0.11 |
ENST00000367242.3
|
TMEM183A
|
transmembrane protein 183A |
| chr3_+_169684553 | 0.11 |
ENST00000337002.4
ENST00000480708.1 |
SEC62
|
SEC62 homolog (S. cerevisiae) |
| chr16_+_22217577 | 0.11 |
ENST00000263026.5
|
EEF2K
|
eukaryotic elongation factor-2 kinase |
| chr19_+_10527449 | 0.11 |
ENST00000592685.1
ENST00000380702.2 |
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr11_-_113746212 | 0.11 |
ENST00000537642.1
ENST00000537706.1 ENST00000544750.1 ENST00000260188.5 ENST00000540925.1 |
USP28
|
ubiquitin specific peptidase 28 |
| chr7_+_128379346 | 0.11 |
ENST00000535011.2
ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU
|
calumenin |
| chr21_-_46237883 | 0.11 |
ENST00000397893.3
|
SUMO3
|
small ubiquitin-like modifier 3 |
| chr4_+_75311019 | 0.11 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr7_-_158622210 | 0.11 |
ENST00000251527.5
|
ESYT2
|
extended synaptotagmin-like protein 2 |
| chr14_-_106092403 | 0.11 |
ENST00000390543.2
|
IGHG4
|
immunoglobulin heavy constant gamma 4 (G4m marker) |
| chr8_-_124408652 | 0.11 |
ENST00000287394.5
|
ATAD2
|
ATPase family, AAA domain containing 2 |
| chr6_-_31865452 | 0.11 |
ENST00000375530.4
ENST00000375537.4 |
EHMT2
|
euchromatic histone-lysine N-methyltransferase 2 |
| chr1_-_226129083 | 0.11 |
ENST00000420304.2
|
LEFTY2
|
left-right determination factor 2 |
| chr17_+_30348024 | 0.11 |
ENST00000327564.7
ENST00000584368.1 ENST00000394713.3 ENST00000341671.7 |
LRRC37B
|
leucine rich repeat containing 37B |
| chr10_+_89622870 | 0.10 |
ENST00000371953.3
|
PTEN
|
phosphatase and tensin homolog |
| chr8_+_22422749 | 0.10 |
ENST00000523900.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr10_+_93558069 | 0.10 |
ENST00000371627.4
|
TNKS2
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 |
| chr16_+_2570340 | 0.10 |
ENST00000568263.1
ENST00000293971.6 ENST00000302956.4 ENST00000413459.3 ENST00000566706.1 ENST00000569879.1 |
AMDHD2
|
amidohydrolase domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.6 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 0.6 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.1 | 0.6 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.1 | 0.3 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 0.1 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.1 | 0.6 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.1 | 1.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.3 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.1 | 0.7 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 0.3 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.6 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.7 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.3 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.5 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.9 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 1.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 0.5 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.6 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 1.0 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.4 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.1 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.5 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.0 | 0.3 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.5 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.2 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.1 | GO:0039019 | regulation of polarized epithelial cell differentiation(GO:0030860) pronephric nephron development(GO:0039019) |
| 0.0 | 0.1 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.0 | 0.0 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 0.0 | 0.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.1 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.0 | 0.4 | GO:0003085 | negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.0 | 0.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.2 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.0 | 0.1 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.6 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.2 | GO:1903298 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) intrinsic apoptotic signaling pathway in response to hypoxia(GO:1990144) |
| 0.0 | 0.2 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.2 | GO:0006489 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0003193 | pulmonary valve formation(GO:0003193) visceral motor neuron differentiation(GO:0021524) foramen ovale closure(GO:0035922) |
| 0.0 | 0.1 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.2 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.0 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.0 | 0.3 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.1 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.0 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.0 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.1 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.2 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 0.6 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 1.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.9 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.2 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 0.6 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.8 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.6 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.3 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.0 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.2 | 0.6 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.1 | 0.6 | GO:0010465 | neurotrophin p75 receptor binding(GO:0005166) nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 1.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.9 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 1.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.3 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.1 | 0.5 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.2 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 0.4 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.5 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.3 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.2 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.0 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.0 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.6 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.2 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.6 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.6 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.2 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |