Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for BHLHE22_BHLHA15_BHLHE23
Z-value: 0.65
Transcription factors associated with BHLHE22_BHLHA15_BHLHE23
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
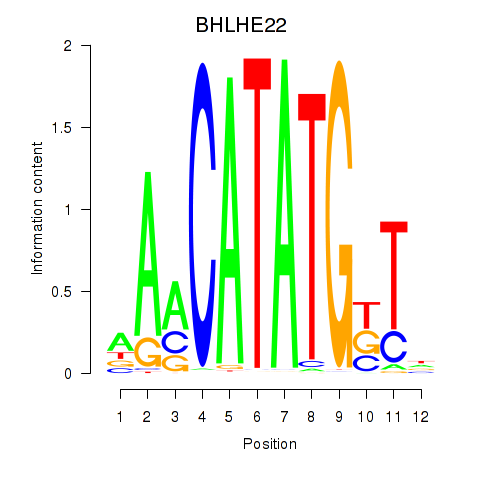
BHLHE22
|
ENSG00000180828.1 | basic helix-loop-helix family member e22 |
|
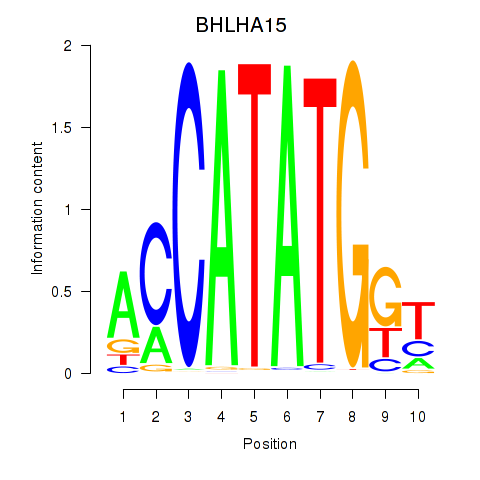
BHLHA15
|
ENSG00000180535.3 | basic helix-loop-helix family member a15 |
|
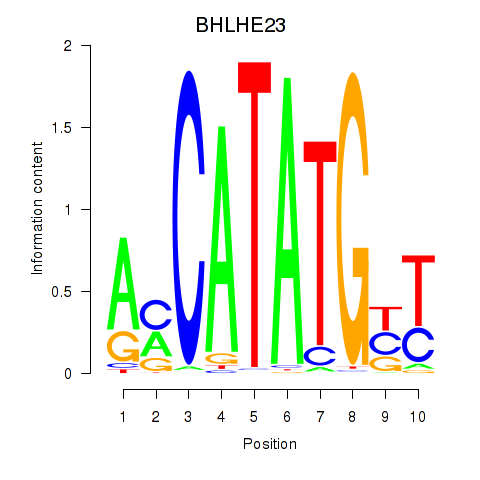
BHLHE23
|
ENSG00000125533.4 | basic helix-loop-helix family member e23 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BHLHE22 | hg19_v2_chr8_+_65492756_65492814 | 0.11 | 6.0e-01 | Click! |
| BHLHE23 | hg19_v2_chr20_-_61638313_61638387 | 0.10 | 6.4e-01 | Click! |
Activity profile of BHLHE22_BHLHA15_BHLHE23 motif
Sorted Z-values of BHLHE22_BHLHA15_BHLHE23 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_111312622 | 2.72 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr10_+_5135981 | 2.02 |
ENST00000380554.3
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr3_-_18480260 | 1.83 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr7_-_27205136 | 1.82 |
ENST00000396345.1
ENST00000343483.6 |
HOXA9
|
homeobox A9 |
| chr1_+_47489240 | 1.72 |
ENST00000371901.3
|
CYP4X1
|
cytochrome P450, family 4, subfamily X, polypeptide 1 |
| chr19_-_11450249 | 1.64 |
ENST00000222120.3
|
RAB3D
|
RAB3D, member RAS oncogene family |
| chr2_-_165424973 | 1.64 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr1_+_61330931 | 1.60 |
ENST00000371191.1
|
NFIA
|
nuclear factor I/A |
| chr1_+_244214577 | 1.29 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr20_+_57226841 | 1.19 |
ENST00000358029.4
ENST00000361830.3 |
STX16
|
syntaxin 16 |
| chr2_-_183387430 | 1.11 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr3_-_114477962 | 1.09 |
ENST00000471418.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr2_-_188378368 | 1.04 |
ENST00000392365.1
ENST00000435414.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr3_-_114477787 | 1.02 |
ENST00000464560.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr7_+_116502605 | 0.94 |
ENST00000458284.2
ENST00000490693.1 |
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr2_-_183387064 | 0.86 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr15_+_65843130 | 0.81 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr9_+_2158485 | 0.72 |
ENST00000417599.1
ENST00000382185.1 ENST00000382183.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_-_197300194 | 0.72 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr9_+_2158443 | 0.69 |
ENST00000302401.3
ENST00000324954.5 ENST00000423555.1 ENST00000382186.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr15_-_55700216 | 0.69 |
ENST00000569205.1
|
CCPG1
|
cell cycle progression 1 |
| chr5_+_175299743 | 0.68 |
ENST00000502265.1
|
CPLX2
|
complexin 2 |
| chr13_+_28813645 | 0.66 |
ENST00000282391.5
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr8_-_38239732 | 0.63 |
ENST00000534155.1
ENST00000433384.2 ENST00000317025.8 ENST00000316985.3 |
WHSC1L1
|
Wolf-Hirschhorn syndrome candidate 1-like 1 |
| chr4_+_144354644 | 0.62 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr3_-_178984759 | 0.61 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr15_+_40650408 | 0.61 |
ENST00000267889.3
|
DISP2
|
dispatched homolog 2 (Drosophila) |
| chr14_+_22675388 | 0.60 |
ENST00000390461.2
|
TRAV34
|
T cell receptor alpha variable 34 |
| chr7_+_116502527 | 0.57 |
ENST00000361183.3
|
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr4_-_159094194 | 0.52 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr7_-_38403077 | 0.52 |
ENST00000426402.2
|
TRGV2
|
T cell receptor gamma variable 2 |
| chr15_-_55700522 | 0.51 |
ENST00000564092.1
ENST00000310958.6 |
CCPG1
|
cell cycle progression 1 |
| chr19_+_18208603 | 0.51 |
ENST00000262811.6
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr15_-_65503801 | 0.48 |
ENST00000261883.4
|
CILP
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr4_-_141075330 | 0.47 |
ENST00000509479.2
|
MAML3
|
mastermind-like 3 (Drosophila) |
| chr5_-_114632307 | 0.47 |
ENST00000506442.1
ENST00000379611.5 |
CCDC112
|
coiled-coil domain containing 112 |
| chr19_-_37019562 | 0.41 |
ENST00000523638.1
|
ZNF260
|
zinc finger protein 260 |
| chrX_+_70798261 | 0.39 |
ENST00000373696.3
|
ACRC
|
acidic repeat containing |
| chr13_+_49551020 | 0.38 |
ENST00000541916.1
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr2_-_183387283 | 0.37 |
ENST00000435564.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chrX_-_19817869 | 0.37 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr4_-_74088800 | 0.37 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr12_-_102872317 | 0.35 |
ENST00000424202.2
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr4_+_25915896 | 0.35 |
ENST00000514384.1
|
SMIM20
|
small integral membrane protein 20 |
| chr1_+_159272111 | 0.34 |
ENST00000368114.1
|
FCER1A
|
Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide |
| chr2_-_166930131 | 0.33 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr10_-_48416849 | 0.32 |
ENST00000249598.1
|
GDF2
|
growth differentiation factor 2 |
| chr12_-_30887948 | 0.32 |
ENST00000433722.2
|
CAPRIN2
|
caprin family member 2 |
| chr6_+_97010424 | 0.31 |
ENST00000541107.1
ENST00000450218.1 ENST00000326771.2 |
FHL5
|
four and a half LIM domains 5 |
| chr8_-_135522425 | 0.31 |
ENST00000521673.1
|
ZFAT
|
zinc finger and AT hook domain containing |
| chr15_-_55700457 | 0.30 |
ENST00000442196.3
ENST00000563171.1 ENST00000425574.3 |
CCPG1
|
cell cycle progression 1 |
| chr17_+_7211656 | 0.29 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr6_-_119031228 | 0.28 |
ENST00000392500.3
ENST00000368488.5 ENST00000434604.1 |
CEP85L
|
centrosomal protein 85kDa-like |
| chr1_-_186344802 | 0.26 |
ENST00000451586.1
|
TPR
|
translocated promoter region, nuclear basket protein |
| chr17_+_60758814 | 0.25 |
ENST00000579432.1
ENST00000446119.2 |
MRC2
|
mannose receptor, C type 2 |
| chr10_-_94257512 | 0.24 |
ENST00000371581.5
|
IDE
|
insulin-degrading enzyme |
| chr8_+_21823726 | 0.23 |
ENST00000433566.4
|
XPO7
|
exportin 7 |
| chr9_+_18474163 | 0.21 |
ENST00000380566.4
|
ADAMTSL1
|
ADAMTS-like 1 |
| chr20_-_34117447 | 0.20 |
ENST00000246199.2
ENST00000424444.1 ENST00000374345.4 ENST00000444723.1 |
C20orf173
|
chromosome 20 open reading frame 173 |
| chr4_+_2813946 | 0.19 |
ENST00000442312.2
|
SH3BP2
|
SH3-domain binding protein 2 |
| chr3_+_46924829 | 0.19 |
ENST00000313049.5
|
PTH1R
|
parathyroid hormone 1 receptor |
| chr6_-_169364429 | 0.19 |
ENST00000444586.1
|
RP3-495K2.3
|
RP3-495K2.3 |
| chr19_-_54876558 | 0.19 |
ENST00000391742.2
ENST00000434277.2 |
LAIR1
|
leukocyte-associated immunoglobulin-like receptor 1 |
| chrX_+_107334895 | 0.18 |
ENST00000372232.3
ENST00000345734.3 ENST00000372254.3 |
ATG4A
|
autophagy related 4A, cysteine peptidase |
| chr4_-_147043058 | 0.18 |
ENST00000512063.1
ENST00000507726.1 |
RP11-6L6.3
|
long intergenic non-protein coding RNA 1095 |
| chr17_-_39597173 | 0.17 |
ENST00000246646.3
|
KRT38
|
keratin 38 |
| chr18_-_52989525 | 0.17 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr3_+_97868170 | 0.16 |
ENST00000437310.1
|
OR5H14
|
olfactory receptor, family 5, subfamily H, member 14 |
| chr2_+_234637754 | 0.16 |
ENST00000482026.1
ENST00000609767.1 |
UGT1A3
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr1_+_95975672 | 0.16 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr6_-_112115074 | 0.16 |
ENST00000368667.2
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr15_-_63448973 | 0.16 |
ENST00000462430.1
|
RPS27L
|
ribosomal protein S27-like |
| chr2_+_181845843 | 0.15 |
ENST00000602710.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr7_+_148287657 | 0.15 |
ENST00000307003.2
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chr14_-_69864993 | 0.14 |
ENST00000555373.1
|
ERH
|
enhancer of rudimentary homolog (Drosophila) |
| chr6_-_29324054 | 0.14 |
ENST00000543825.1
|
OR5V1
|
olfactory receptor, family 5, subfamily V, member 1 |
| chr3_-_108672609 | 0.13 |
ENST00000393963.3
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr14_+_77292715 | 0.13 |
ENST00000393774.3
ENST00000555189.1 ENST00000450042.2 |
C14orf166B
|
chromosome 14 open reading frame 166B |
| chr8_+_67782984 | 0.13 |
ENST00000396592.3
ENST00000422365.2 ENST00000492775.1 |
MCMDC2
|
minichromosome maintenance domain containing 2 |
| chr2_+_210444748 | 0.13 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr6_+_73076432 | 0.13 |
ENST00000414192.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr7_-_112758665 | 0.12 |
ENST00000397764.3
|
LINC00998
|
long intergenic non-protein coding RNA 998 |
| chr14_+_27342334 | 0.11 |
ENST00000548170.1
ENST00000552926.1 |
RP11-384J4.1
|
RP11-384J4.1 |
| chr13_+_111766897 | 0.11 |
ENST00000317133.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chrX_+_105855160 | 0.11 |
ENST00000372544.2
ENST00000372548.4 |
CXorf57
|
chromosome X open reading frame 57 |
| chr3_-_33700933 | 0.11 |
ENST00000480013.1
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr17_-_47286729 | 0.11 |
ENST00000300406.2
ENST00000511277.1 ENST00000511673.1 |
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr15_+_85144217 | 0.10 |
ENST00000540936.1
ENST00000448803.2 ENST00000546275.1 ENST00000546148.1 ENST00000442073.3 ENST00000334141.3 ENST00000358472.3 ENST00000502939.2 ENST00000379358.3 ENST00000327179.6 |
ZSCAN2
|
zinc finger and SCAN domain containing 2 |
| chr1_-_44818599 | 0.10 |
ENST00000537474.1
|
ERI3
|
ERI1 exoribonuclease family member 3 |
| chr10_-_15902449 | 0.10 |
ENST00000277632.3
|
FAM188A
|
family with sequence similarity 188, member A |
| chr3_-_33700589 | 0.09 |
ENST00000461133.3
ENST00000496954.2 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr3_-_108672742 | 0.09 |
ENST00000261047.3
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr8_+_67976593 | 0.09 |
ENST00000262210.5
ENST00000412460.1 |
CSPP1
|
centrosome and spindle pole associated protein 1 |
| chr7_+_114562909 | 0.09 |
ENST00000423503.1
ENST00000427207.1 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr3_+_97887544 | 0.09 |
ENST00000356526.2
|
OR5H15
|
olfactory receptor, family 5, subfamily H, member 15 |
| chr6_-_117150198 | 0.09 |
ENST00000310357.3
ENST00000368549.3 ENST00000530250.1 |
GPRC6A
|
G protein-coupled receptor, family C, group 6, member A |
| chr16_+_14726672 | 0.08 |
ENST00000261658.2
ENST00000563971.1 |
BFAR
|
bifunctional apoptosis regulator |
| chr7_-_41742697 | 0.08 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr11_-_62359027 | 0.08 |
ENST00000494385.1
ENST00000308436.7 |
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr3_+_137717571 | 0.08 |
ENST00000343735.4
|
CLDN18
|
claudin 18 |
| chr7_-_97881429 | 0.07 |
ENST00000420697.1
ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr12_+_50794730 | 0.07 |
ENST00000523389.1
ENST00000518561.1 ENST00000347328.5 ENST00000550260.1 |
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr5_+_156607829 | 0.06 |
ENST00000422843.3
|
ITK
|
IL2-inducible T-cell kinase |
| chr2_-_227050079 | 0.06 |
ENST00000423838.1
|
AC068138.1
|
AC068138.1 |
| chr14_+_24540046 | 0.06 |
ENST00000397016.2
ENST00000537691.1 ENST00000560356.1 ENST00000558450.1 |
CPNE6
|
copine VI (neuronal) |
| chr21_-_36421401 | 0.06 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr4_+_2814011 | 0.06 |
ENST00000502260.1
ENST00000435136.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr16_-_52640834 | 0.05 |
ENST00000510238.3
|
CASC16
|
cancer susceptibility candidate 16 (non-protein coding) |
| chr12_-_91573316 | 0.05 |
ENST00000393155.1
|
DCN
|
decorin |
| chr1_+_12524965 | 0.05 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr7_-_101212244 | 0.04 |
ENST00000451953.1
ENST00000434537.1 ENST00000437900.1 |
LINC01007
|
long intergenic non-protein coding RNA 1007 |
| chrX_+_120181457 | 0.04 |
ENST00000328078.1
|
GLUD2
|
glutamate dehydrogenase 2 |
| chrX_+_107288239 | 0.03 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr3_+_113251143 | 0.03 |
ENST00000264852.4
ENST00000393830.3 |
SIDT1
|
SID1 transmembrane family, member 1 |
| chr3_-_53164423 | 0.03 |
ENST00000467048.1
ENST00000394738.3 ENST00000296292.3 |
RFT1
|
RFT1 homolog (S. cerevisiae) |
| chr3_+_112930306 | 0.03 |
ENST00000495514.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr19_-_52227221 | 0.03 |
ENST00000222115.1
ENST00000540069.2 |
HAS1
|
hyaluronan synthase 1 |
| chr7_-_99573677 | 0.03 |
ENST00000292401.4
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr3_+_137728842 | 0.03 |
ENST00000183605.5
|
CLDN18
|
claudin 18 |
| chr4_+_69313145 | 0.03 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr3_+_119298429 | 0.02 |
ENST00000478927.1
|
ADPRH
|
ADP-ribosylarginine hydrolase |
| chr11_+_66008698 | 0.02 |
ENST00000531597.1
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr8_+_62747349 | 0.02 |
ENST00000517953.1
ENST00000520097.1 ENST00000519766.1 |
RP11-705O24.1
|
RP11-705O24.1 |
| chr9_+_12775011 | 0.02 |
ENST00000319264.3
|
LURAP1L
|
leucine rich adaptor protein 1-like |
| chr6_+_37400974 | 0.02 |
ENST00000455891.1
ENST00000373451.4 |
CMTR1
|
cap methyltransferase 1 |
| chr1_-_157670647 | 0.02 |
ENST00000368184.3
|
FCRL3
|
Fc receptor-like 3 |
| chr11_-_44972476 | 0.02 |
ENST00000527685.1
ENST00000308212.5 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr12_-_95397442 | 0.02 |
ENST00000547157.1
ENST00000547986.1 ENST00000327772.2 |
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chrX_-_15402498 | 0.01 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr8_-_67976509 | 0.01 |
ENST00000518747.1
|
COPS5
|
COP9 signalosome subunit 5 |
| chr4_+_154178520 | 0.00 |
ENST00000433687.1
|
TRIM2
|
tripartite motif containing 2 |
| chr2_-_32490859 | 0.00 |
ENST00000404025.2
|
NLRC4
|
NLR family, CARD domain containing 4 |
| chr12_-_108714412 | 0.00 |
ENST00000412676.1
ENST00000550573.1 |
CMKLR1
|
chemokine-like receptor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BHLHE22_BHLHA15_BHLHE23
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.3 | 1.6 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.3 | 1.5 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.7 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 1.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 2.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.3 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.1 | 1.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 1.8 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.4 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.3 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 | 0.2 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 0.4 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 1.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.7 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 1.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.5 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 1.5 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 2.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.5 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.2 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 1.6 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.5 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.3 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.6 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 1.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.7 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 1.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 1.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.4 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 1.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 1.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.7 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0047718 | androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.3 | 2.3 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 0.7 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 1.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 1.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) IgE binding(GO:0019863) |
| 0.0 | 0.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.2 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 1.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 1.4 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.6 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.6 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 1.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.6 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.0 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 1.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |