Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for BHLHE40
Z-value: 0.57
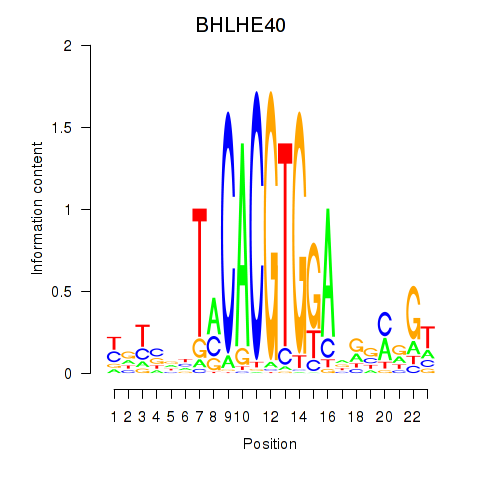
Transcription factors associated with BHLHE40
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BHLHE40
|
ENSG00000134107.4 | basic helix-loop-helix family member e40 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BHLHE40 | hg19_v2_chr3_+_5020801_5020952 | 0.07 | 7.4e-01 | Click! |
Activity profile of BHLHE40 motif
Sorted Z-values of BHLHE40 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_6019552 | 1.97 |
ENST00000379977.3
ENST00000397251.3 ENST00000397248.2 |
IL15RA
|
interleukin 15 receptor, alpha |
| chr9_-_123691047 | 1.55 |
ENST00000373887.3
|
TRAF1
|
TNF receptor-associated factor 1 |
| chr10_-_6019455 | 1.26 |
ENST00000530685.1
ENST00000397255.3 ENST00000379971.1 ENST00000528354.1 ENST00000397250.2 ENST00000429135.2 |
IL15RA
|
interleukin 15 receptor, alpha |
| chr2_+_127413481 | 1.12 |
ENST00000259254.4
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr12_+_66217911 | 1.07 |
ENST00000403681.2
|
HMGA2
|
high mobility group AT-hook 2 |
| chr2_-_136743169 | 1.06 |
ENST00000264161.4
|
DARS
|
aspartyl-tRNA synthetase |
| chr1_-_165325939 | 1.01 |
ENST00000342310.3
|
LMX1A
|
LIM homeobox transcription factor 1, alpha |
| chr16_-_56701933 | 1.01 |
ENST00000568675.1
ENST00000569500.1 ENST00000444837.2 ENST00000379811.3 |
MT1G
|
metallothionein 1G |
| chr2_-_136743039 | 0.99 |
ENST00000537273.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr22_+_41697520 | 0.93 |
ENST00000352645.4
|
ZC3H7B
|
zinc finger CCCH-type containing 7B |
| chr8_+_32405785 | 0.82 |
ENST00000287842.3
|
NRG1
|
neuregulin 1 |
| chr2_-_10588630 | 0.79 |
ENST00000234111.4
|
ODC1
|
ornithine decarboxylase 1 |
| chr8_+_32405728 | 0.79 |
ENST00000523079.1
ENST00000338921.4 ENST00000356819.4 ENST00000287845.5 ENST00000341377.5 |
NRG1
|
neuregulin 1 |
| chr9_-_100881466 | 0.78 |
ENST00000341469.2
ENST00000342043.3 ENST00000375098.3 |
TRIM14
|
tripartite motif containing 14 |
| chr2_-_10587897 | 0.74 |
ENST00000405333.1
ENST00000443218.1 |
ODC1
|
ornithine decarboxylase 1 |
| chr22_-_50964558 | 0.72 |
ENST00000535425.1
ENST00000439934.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr19_-_55652290 | 0.65 |
ENST00000589745.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr12_-_58165870 | 0.54 |
ENST00000257848.7
|
METTL1
|
methyltransferase like 1 |
| chr8_+_99129513 | 0.54 |
ENST00000522319.1
ENST00000401707.2 |
POP1
|
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr11_+_6624970 | 0.54 |
ENST00000420936.2
ENST00000528995.1 |
ILK
|
integrin-linked kinase |
| chr1_-_231376836 | 0.53 |
ENST00000451322.1
|
C1orf131
|
chromosome 1 open reading frame 131 |
| chr3_+_133293278 | 0.49 |
ENST00000508481.1
ENST00000420115.2 ENST00000504867.1 ENST00000507408.1 ENST00000511392.1 ENST00000515421.1 |
CDV3
|
CDV3 homolog (mouse) |
| chr11_+_6625046 | 0.47 |
ENST00000396751.2
|
ILK
|
integrin-linked kinase |
| chr14_+_77924373 | 0.47 |
ENST00000216479.3
ENST00000535854.2 ENST00000555517.1 |
AHSA1
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast) |
| chr11_+_6624955 | 0.47 |
ENST00000299421.4
ENST00000537806.1 |
ILK
|
integrin-linked kinase |
| chr6_-_5260963 | 0.46 |
ENST00000464010.1
ENST00000468929.1 ENST00000480566.1 |
LYRM4
|
LYR motif containing 4 |
| chr1_-_171621815 | 0.45 |
ENST00000037502.6
|
MYOC
|
myocilin, trabecular meshwork inducible glucocorticoid response |
| chr6_-_5261141 | 0.43 |
ENST00000330636.4
ENST00000500576.2 |
LYRM4
|
LYR motif containing 4 |
| chr6_-_36842784 | 0.42 |
ENST00000373699.5
|
PPIL1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr17_+_33307503 | 0.42 |
ENST00000378526.4
ENST00000585941.1 ENST00000262327.5 ENST00000592690.1 ENST00000585740.1 |
LIG3
|
ligase III, DNA, ATP-dependent |
| chr7_-_100493744 | 0.42 |
ENST00000428317.1
ENST00000441605.1 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr14_+_73393040 | 0.40 |
ENST00000358377.2
ENST00000353777.3 ENST00000394234.2 ENST00000509153.1 ENST00000555042.1 |
DCAF4
|
DDB1 and CUL4 associated factor 4 |
| chr17_-_10600818 | 0.40 |
ENST00000577427.1
ENST00000255390.5 |
SCO1
|
SCO1 cytochrome c oxidase assembly protein |
| chr19_+_35645618 | 0.40 |
ENST00000392218.2
ENST00000543307.1 ENST00000392219.2 ENST00000541435.2 ENST00000590686.1 ENST00000342879.3 ENST00000588699.1 |
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr7_+_28725585 | 0.39 |
ENST00000396298.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr5_+_172571445 | 0.35 |
ENST00000231668.9
ENST00000351486.5 ENST00000352523.6 ENST00000393770.4 |
BNIP1
|
BCL2/adenovirus E1B 19kDa interacting protein 1 |
| chr1_+_213123915 | 0.34 |
ENST00000366968.4
ENST00000490792.1 |
VASH2
|
vasohibin 2 |
| chr8_+_125985531 | 0.34 |
ENST00000319286.5
|
ZNF572
|
zinc finger protein 572 |
| chr9_+_130565147 | 0.33 |
ENST00000373247.2
ENST00000373245.1 ENST00000393706.2 ENST00000373228.1 |
FPGS
|
folylpolyglutamate synthase |
| chr19_-_5719860 | 0.31 |
ENST00000590729.1
|
LONP1
|
lon peptidase 1, mitochondrial |
| chr6_+_70576457 | 0.31 |
ENST00000322773.4
|
COL19A1
|
collagen, type XIX, alpha 1 |
| chr19_+_35645817 | 0.30 |
ENST00000423817.3
|
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr10_-_35379241 | 0.30 |
ENST00000374748.1
ENST00000374749.3 |
CUL2
|
cullin 2 |
| chr18_+_580367 | 0.29 |
ENST00000327228.3
|
CETN1
|
centrin, EF-hand protein, 1 |
| chr3_-_21792838 | 0.29 |
ENST00000281523.2
|
ZNF385D
|
zinc finger protein 385D |
| chr7_-_151574191 | 0.27 |
ENST00000287878.4
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr14_+_93389425 | 0.26 |
ENST00000216492.5
ENST00000334654.4 |
CHGA
|
chromogranin A (parathyroid secretory protein 1) |
| chr12_-_6677422 | 0.26 |
ENST00000382421.3
ENST00000545200.1 ENST00000399466.2 ENST00000536124.1 ENST00000540228.1 ENST00000542867.1 ENST00000545492.1 ENST00000322166.5 ENST00000545915.1 |
NOP2
|
NOP2 nucleolar protein |
| chr12_-_48298785 | 0.26 |
ENST00000550325.1
ENST00000546653.1 ENST00000549336.1 ENST00000535672.1 ENST00000229022.3 ENST00000548664.1 |
VDR
|
vitamin D (1,25- dihydroxyvitamin D3) receptor |
| chr6_+_13615554 | 0.24 |
ENST00000451315.2
|
NOL7
|
nucleolar protein 7, 27kDa |
| chr9_+_706842 | 0.24 |
ENST00000382293.3
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr8_+_55047763 | 0.24 |
ENST00000260102.4
ENST00000519831.1 |
MRPL15
|
mitochondrial ribosomal protein L15 |
| chr22_-_37505449 | 0.24 |
ENST00000406725.1
|
TMPRSS6
|
transmembrane protease, serine 6 |
| chr9_-_95640218 | 0.23 |
ENST00000395506.3
ENST00000375495.3 ENST00000332591.6 |
ZNF484
|
zinc finger protein 484 |
| chr1_-_112046110 | 0.23 |
ENST00000369716.4
|
ADORA3
|
adenosine A3 receptor |
| chr19_-_45909585 | 0.22 |
ENST00000593226.1
ENST00000418234.2 |
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like |
| chrX_+_129040122 | 0.22 |
ENST00000394422.3
ENST00000371051.5 |
UTP14A
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr5_-_114880533 | 0.22 |
ENST00000274457.3
|
FEM1C
|
fem-1 homolog c (C. elegans) |
| chr6_-_136871957 | 0.22 |
ENST00000354570.3
|
MAP7
|
microtubule-associated protein 7 |
| chr11_+_60609537 | 0.21 |
ENST00000227520.5
|
CCDC86
|
coiled-coil domain containing 86 |
| chr17_-_17109579 | 0.21 |
ENST00000321560.3
|
PLD6
|
phospholipase D family, member 6 |
| chr1_-_112046289 | 0.21 |
ENST00000241356.4
|
ADORA3
|
adenosine A3 receptor |
| chr8_+_109455845 | 0.21 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr2_+_201676908 | 0.21 |
ENST00000409226.1
ENST00000452790.2 |
BZW1
|
basic leucine zipper and W2 domains 1 |
| chr1_+_48688357 | 0.20 |
ENST00000533824.1
ENST00000438567.2 ENST00000236495.5 ENST00000420136.2 |
SLC5A9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr5_+_89770696 | 0.20 |
ENST00000504930.1
ENST00000514483.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chrX_+_129040094 | 0.19 |
ENST00000425117.2
|
UTP14A
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr22_-_50523760 | 0.19 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr10_-_35379524 | 0.19 |
ENST00000374751.3
ENST00000374742.1 ENST00000602371.1 |
CUL2
|
cullin 2 |
| chr1_-_86174065 | 0.18 |
ENST00000370574.3
ENST00000431532.2 |
ZNHIT6
|
zinc finger, HIT-type containing 6 |
| chr2_+_183989157 | 0.17 |
ENST00000541912.1
|
NUP35
|
nucleoporin 35kDa |
| chr1_-_43638168 | 0.17 |
ENST00000431635.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chrX_+_54556633 | 0.17 |
ENST00000336470.4
ENST00000360845.2 |
GNL3L
|
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr5_+_89770664 | 0.17 |
ENST00000503973.1
ENST00000399107.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr1_+_207070775 | 0.16 |
ENST00000391929.3
ENST00000294984.2 ENST00000367093.3 |
IL24
|
interleukin 24 |
| chr4_+_119200215 | 0.16 |
ENST00000602573.1
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr8_-_11710979 | 0.16 |
ENST00000415599.2
|
CTSB
|
cathepsin B |
| chr22_-_30987849 | 0.16 |
ENST00000402284.3
ENST00000354694.7 |
PES1
|
pescadillo ribosomal biogenesis factor 1 |
| chr11_-_6624801 | 0.16 |
ENST00000534343.1
ENST00000254605.6 |
RRP8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr12_-_55042140 | 0.15 |
ENST00000293371.6
ENST00000456047.2 |
DCD
|
dermcidin |
| chr2_+_183989083 | 0.15 |
ENST00000295119.4
|
NUP35
|
nucleoporin 35kDa |
| chr6_-_151773232 | 0.14 |
ENST00000444024.1
ENST00000367303.4 |
RMND1
|
required for meiotic nuclear division 1 homolog (S. cerevisiae) |
| chr16_+_29911864 | 0.14 |
ENST00000308748.5
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr4_+_119199864 | 0.14 |
ENST00000602414.1
ENST00000602520.1 |
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr1_-_43637915 | 0.14 |
ENST00000236051.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr2_+_207630081 | 0.14 |
ENST00000236980.6
ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2
|
FAST kinase domains 2 |
| chr20_+_23331373 | 0.14 |
ENST00000254998.2
|
NXT1
|
NTF2-like export factor 1 |
| chr10_-_21435488 | 0.14 |
ENST00000534331.1
ENST00000529198.1 ENST00000377118.4 |
C10orf113
|
chromosome 10 open reading frame 113 |
| chr14_+_22694606 | 0.13 |
ENST00000390463.3
|
TRAV36DV7
|
T cell receptor alpha variable 36/delta variable 7 |
| chr21_-_45079341 | 0.13 |
ENST00000443485.1
ENST00000291560.2 |
HSF2BP
|
heat shock transcription factor 2 binding protein |
| chr17_+_40811283 | 0.13 |
ENST00000251412.7
|
TUBG2
|
tubulin, gamma 2 |
| chr22_-_30987837 | 0.13 |
ENST00000335214.6
|
PES1
|
pescadillo ribosomal biogenesis factor 1 |
| chr9_+_37120536 | 0.12 |
ENST00000336755.5
ENST00000534928.1 ENST00000322831.6 |
ZCCHC7
|
zinc finger, CCHC domain containing 7 |
| chr17_-_7193711 | 0.12 |
ENST00000571464.1
|
YBX2
|
Y box binding protein 2 |
| chr19_-_3786253 | 0.12 |
ENST00000585778.1
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr16_-_20362147 | 0.11 |
ENST00000396142.2
|
UMOD
|
uromodulin |
| chr4_+_119199904 | 0.11 |
ENST00000602483.1
ENST00000602819.1 |
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr3_+_174577070 | 0.11 |
ENST00000454872.1
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2 |
| chr6_+_36410762 | 0.11 |
ENST00000483557.1
ENST00000498267.1 ENST00000544295.1 ENST00000449081.2 ENST00000536244.1 ENST00000460983.1 |
KCTD20
|
potassium channel tetramerization domain containing 20 |
| chr1_+_213123976 | 0.10 |
ENST00000366965.2
ENST00000366967.2 |
VASH2
|
vasohibin 2 |
| chr17_+_15635561 | 0.10 |
ENST00000584301.1
ENST00000580596.1 ENST00000464963.1 ENST00000437605.2 ENST00000579428.1 |
TBC1D26
|
TBC1 domain family, member 26 |
| chr8_+_26240414 | 0.10 |
ENST00000380629.2
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr4_-_57301748 | 0.10 |
ENST00000264220.2
|
PPAT
|
phosphoribosyl pyrophosphate amidotransferase |
| chr22_-_43411106 | 0.10 |
ENST00000453643.1
ENST00000263246.3 ENST00000337959.4 |
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr19_-_45953983 | 0.09 |
ENST00000592083.1
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr1_-_207095324 | 0.09 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr4_+_57301896 | 0.09 |
ENST00000514888.1
ENST00000264221.2 ENST00000505164.1 |
PAICS
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr17_-_74733404 | 0.08 |
ENST00000508921.3
ENST00000583836.1 ENST00000358156.6 ENST00000392485.2 ENST00000359995.5 |
SRSF2
|
serine/arginine-rich splicing factor 2 |
| chr3_-_157217328 | 0.08 |
ENST00000392832.2
ENST00000543418.1 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr15_-_35088340 | 0.08 |
ENST00000290378.4
|
ACTC1
|
actin, alpha, cardiac muscle 1 |
| chr1_+_118472343 | 0.08 |
ENST00000369441.3
ENST00000349139.5 |
WDR3
|
WD repeat domain 3 |
| chr4_+_41983713 | 0.07 |
ENST00000333141.5
|
DCAF4L1
|
DDB1 and CUL4 associated factor 4-like 1 |
| chr3_+_99536663 | 0.07 |
ENST00000421999.2
ENST00000463526.1 |
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr2_+_103236004 | 0.07 |
ENST00000233969.2
|
SLC9A2
|
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr12_+_65004292 | 0.07 |
ENST00000542104.1
ENST00000336061.2 |
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr22_-_38245304 | 0.07 |
ENST00000609454.1
|
ANKRD54
|
ankyrin repeat domain 54 |
| chr17_+_36908984 | 0.06 |
ENST00000225426.4
ENST00000579088.1 |
PSMB3
|
proteasome (prosome, macropain) subunit, beta type, 3 |
| chr1_-_231376867 | 0.06 |
ENST00000366649.2
ENST00000318906.2 ENST00000366651.3 |
C1orf131
|
chromosome 1 open reading frame 131 |
| chrX_+_23685653 | 0.06 |
ENST00000379331.3
|
PRDX4
|
peroxiredoxin 4 |
| chr6_-_109804412 | 0.06 |
ENST00000230122.3
|
ZBTB24
|
zinc finger and BTB domain containing 24 |
| chr17_-_72772462 | 0.06 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr1_+_21835858 | 0.06 |
ENST00000539907.1
ENST00000540617.1 ENST00000374840.3 |
ALPL
|
alkaline phosphatase, liver/bone/kidney |
| chr13_-_38172863 | 0.06 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr12_-_54673871 | 0.06 |
ENST00000209875.4
|
CBX5
|
chromobox homolog 5 |
| chr4_+_57302297 | 0.05 |
ENST00000399688.3
ENST00000512576.1 |
PAICS
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr7_-_229557 | 0.05 |
ENST00000514988.1
|
AC145676.2
|
Uncharacterized protein |
| chr11_-_65626753 | 0.05 |
ENST00000526975.1
ENST00000531413.1 |
CFL1
|
cofilin 1 (non-muscle) |
| chr17_+_53046096 | 0.05 |
ENST00000376352.2
ENST00000299341.4 ENST00000405898.1 ENST00000434978.2 ENST00000398391.2 |
STXBP4
|
syntaxin binding protein 4 |
| chr6_+_144164455 | 0.04 |
ENST00000367576.5
|
LTV1
|
LTV1 homolog (S. cerevisiae) |
| chr21_-_30391636 | 0.04 |
ENST00000493196.1
|
RWDD2B
|
RWD domain containing 2B |
| chr19_+_56154913 | 0.04 |
ENST00000270451.5
ENST00000588537.1 |
ZNF581
|
zinc finger protein 581 |
| chr19_-_52133588 | 0.04 |
ENST00000570106.2
|
SIGLEC5
|
sialic acid binding Ig-like lectin 5 |
| chr5_-_71616043 | 0.04 |
ENST00000508863.2
ENST00000522095.1 ENST00000513900.1 ENST00000515404.1 ENST00000457646.4 ENST00000261413.5 |
MRPS27
|
mitochondrial ribosomal protein S27 |
| chr5_-_101834617 | 0.04 |
ENST00000513675.1
ENST00000379807.3 |
SLCO6A1
|
solute carrier organic anion transporter family, member 6A1 |
| chr7_+_141463897 | 0.04 |
ENST00000247879.2
|
TAS2R3
|
taste receptor, type 2, member 3 |
| chr7_+_147830776 | 0.04 |
ENST00000538075.1
|
CNTNAP2
|
contactin associated protein-like 2 |
| chr16_+_19729586 | 0.03 |
ENST00000564186.1
ENST00000541926.1 ENST00000433597.2 |
IQCK
|
IQ motif containing K |
| chr1_+_210001309 | 0.03 |
ENST00000491415.2
|
DIEXF
|
digestive organ expansion factor homolog (zebrafish) |
| chr5_-_101834712 | 0.03 |
ENST00000506729.1
ENST00000389019.3 ENST00000379810.1 |
SLCO6A1
|
solute carrier organic anion transporter family, member 6A1 |
| chr1_+_119957554 | 0.03 |
ENST00000543831.1
ENST00000433745.1 ENST00000369416.3 |
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr4_+_188916918 | 0.02 |
ENST00000509524.1
ENST00000326866.4 |
ZFP42
|
ZFP42 zinc finger protein |
| chrX_-_47518527 | 0.02 |
ENST00000333119.3
|
UXT
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr3_+_111578583 | 0.02 |
ENST00000478922.1
ENST00000477695.1 |
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr2_-_47143160 | 0.02 |
ENST00000409800.1
ENST00000409218.1 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr16_+_29911666 | 0.02 |
ENST00000563177.1
ENST00000483405.1 |
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr14_-_102026643 | 0.01 |
ENST00000555882.1
ENST00000554441.1 ENST00000553729.1 ENST00000557109.1 ENST00000557532.1 ENST00000554694.1 ENST00000554735.1 ENST00000555174.1 ENST00000557661.1 |
DIO3OS
|
DIO3 opposite strand/antisense RNA (head to head) |
| chr6_+_31865552 | 0.01 |
ENST00000469372.1
ENST00000497706.1 |
C2
|
complement component 2 |
| chr8_-_48872686 | 0.01 |
ENST00000314191.2
ENST00000338368.3 |
PRKDC
|
protein kinase, DNA-activated, catalytic polypeptide |
| chr11_-_65626797 | 0.01 |
ENST00000525451.2
|
CFL1
|
cofilin 1 (non-muscle) |
| chr16_+_4897912 | 0.01 |
ENST00000545171.1
|
UBN1
|
ubinuclein 1 |
| chr5_+_94982435 | 0.01 |
ENST00000511684.1
ENST00000380005.4 |
RFESD
|
Rieske (Fe-S) domain containing |
| chr12_+_93861264 | 0.01 |
ENST00000549982.1
ENST00000361630.2 |
MRPL42
|
mitochondrial ribosomal protein L42 |
| chr7_-_155326532 | 0.00 |
ENST00000406197.1
ENST00000321736.5 |
CNPY1
|
canopy FGF signaling regulator 1 |
| chr17_-_8066843 | 0.00 |
ENST00000404970.3
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2) |
| chr19_+_7660716 | 0.00 |
ENST00000160298.4
ENST00000446248.2 |
CAMSAP3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr17_-_61850894 | 0.00 |
ENST00000403162.3
ENST00000582252.1 ENST00000225726.5 |
CCDC47
|
coiled-coil domain containing 47 |
| chr16_+_4897632 | 0.00 |
ENST00000262376.6
|
UBN1
|
ubinuclein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BHLHE40
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.4 | 1.5 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.3 | 1.1 | GO:0031052 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.3 | 2.1 | GO:0038129 | ERBB3 signaling pathway(GO:0038129) |
| 0.2 | 0.7 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 | 0.4 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 0.3 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 0.4 | GO:1901859 | base-excision repair, DNA ligation(GO:0006288) negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.1 | 0.3 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.1 | 0.3 | GO:0061110 | dense core granule biogenesis(GO:0061110) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.3 | GO:1904747 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.1 | 0.2 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.6 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.5 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.1 | 1.0 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.1 | 1.0 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.1 | 1.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 1.5 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.2 | GO:0010635 | regulation of mitochondrial fusion(GO:0010635) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.3 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.8 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.4 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.4 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 1.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 1.6 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.3 | GO:0072584 | caveolin-mediated endocytosis(GO:0072584) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.0 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.0 | 0.3 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.5 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 1.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.5 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 0.5 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.4 | GO:0030430 | host cell cytoplasm(GO:0030430) host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 1.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 1.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.4 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.3 | 1.1 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.2 | 1.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.3 | GO:0070364 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.1 | 0.4 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 1.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.3 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 0.7 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.1 | 0.3 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.1 | 0.3 | GO:0038181 | bile acid receptor activity(GO:0038181) vitamin D response element binding(GO:0070644) |
| 0.0 | 0.6 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.4 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.5 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 1.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 3.2 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.3 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 2.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |