Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
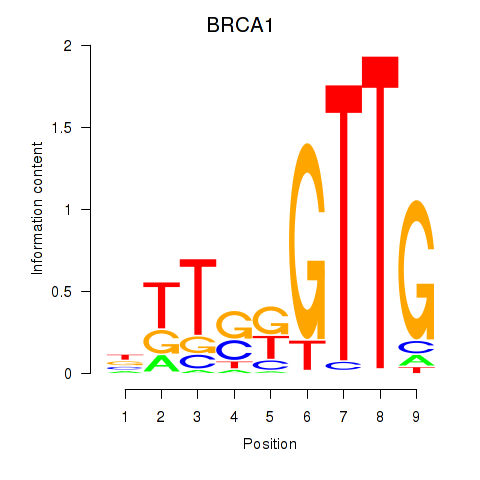
Results for BRCA1
Z-value: 0.39
Transcription factors associated with BRCA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BRCA1
|
ENSG00000012048.15 | BRCA1 DNA repair associated |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BRCA1 | hg19_v2_chr17_-_41277467_41277510 | -0.31 | 1.4e-01 | Click! |
Activity profile of BRCA1 motif
Sorted Z-values of BRCA1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_173020056 | 1.32 |
ENST00000239468.2
ENST00000404377.3 |
TNFSF18
|
tumor necrosis factor (ligand) superfamily, member 18 |
| chr15_+_67430339 | 1.11 |
ENST00000439724.3
|
SMAD3
|
SMAD family member 3 |
| chr2_+_108994633 | 0.80 |
ENST00000409309.3
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr2_+_108994466 | 0.75 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr17_-_41739283 | 0.70 |
ENST00000393661.2
ENST00000318579.4 |
MEOX1
|
mesenchyme homeobox 1 |
| chr17_-_41738931 | 0.69 |
ENST00000329168.3
ENST00000549132.1 |
MEOX1
|
mesenchyme homeobox 1 |
| chr7_+_44646162 | 0.48 |
ENST00000439616.2
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr1_-_160232312 | 0.45 |
ENST00000440682.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr12_-_117318788 | 0.43 |
ENST00000550505.1
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr17_+_74372662 | 0.41 |
ENST00000591651.1
ENST00000545180.1 |
SPHK1
|
sphingosine kinase 1 |
| chr8_+_22022800 | 0.38 |
ENST00000397814.3
|
BMP1
|
bone morphogenetic protein 1 |
| chr10_-_36813162 | 0.38 |
ENST00000440465.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr11_-_119234876 | 0.35 |
ENST00000525735.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr10_+_28822636 | 0.35 |
ENST00000442148.1
ENST00000448193.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr3_-_127455200 | 0.33 |
ENST00000398101.3
|
MGLL
|
monoglyceride lipase |
| chr18_+_19749386 | 0.32 |
ENST00000269216.3
|
GATA6
|
GATA binding protein 6 |
| chr12_-_95510743 | 0.31 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr2_+_203879568 | 0.28 |
ENST00000449802.1
|
NBEAL1
|
neurobeachin-like 1 |
| chr8_-_72459885 | 0.26 |
ENST00000523987.1
|
RP11-1102P16.1
|
Uncharacterized protein |
| chr10_+_104404644 | 0.25 |
ENST00000462202.2
|
TRIM8
|
tripartite motif containing 8 |
| chr18_+_55888767 | 0.24 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr5_+_139027877 | 0.22 |
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5 |
| chr19_+_46850320 | 0.22 |
ENST00000391919.1
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr9_+_135457530 | 0.21 |
ENST00000263610.2
|
BARHL1
|
BarH-like homeobox 1 |
| chr19_+_46850251 | 0.20 |
ENST00000012443.4
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr18_-_23671139 | 0.19 |
ENST00000579061.1
ENST00000542420.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr19_+_55888186 | 0.18 |
ENST00000291934.3
|
TMEM190
|
transmembrane protein 190 |
| chr6_+_42981922 | 0.18 |
ENST00000326974.4
ENST00000244670.8 |
KLHDC3
|
kelch domain containing 3 |
| chr17_-_27230035 | 0.18 |
ENST00000378895.4
ENST00000394901.3 |
DHRS13
|
dehydrogenase/reductase (SDR family) member 13 |
| chr10_-_95504 | 0.18 |
ENST00000447903.2
|
TUBB8
|
tubulin, beta 8 class VIII |
| chr7_+_44646177 | 0.18 |
ENST00000443864.2
ENST00000447398.1 ENST00000449767.1 ENST00000419661.1 |
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr1_+_10271674 | 0.17 |
ENST00000377086.1
|
KIF1B
|
kinesin family member 1B |
| chr6_+_12717892 | 0.17 |
ENST00000379350.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chrX_+_10124977 | 0.17 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr17_+_36584662 | 0.16 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr8_-_81083341 | 0.15 |
ENST00000519303.2
|
TPD52
|
tumor protein D52 |
| chr17_-_60142609 | 0.15 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chr2_-_158182410 | 0.15 |
ENST00000419116.2
ENST00000410096.1 |
ERMN
|
ermin, ERM-like protein |
| chr19_-_48059113 | 0.15 |
ENST00000391901.3
ENST00000314121.4 ENST00000448976.1 |
ZNF541
|
zinc finger protein 541 |
| chr11_-_71823266 | 0.14 |
ENST00000538919.1
ENST00000539395.1 ENST00000542531.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chrX_-_70288234 | 0.13 |
ENST00000276105.3
ENST00000374274.3 |
SNX12
|
sorting nexin 12 |
| chr4_+_619347 | 0.11 |
ENST00000255622.6
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr10_-_95209 | 0.10 |
ENST00000332708.5
ENST00000309812.4 |
TUBB8
|
tubulin, beta 8 class VIII |
| chr19_-_40324767 | 0.10 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr12_-_12491608 | 0.10 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr12_-_117319236 | 0.10 |
ENST00000257572.5
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr13_+_30002846 | 0.09 |
ENST00000542829.1
|
MTUS2
|
microtubule associated tumor suppressor candidate 2 |
| chr19_+_45394477 | 0.09 |
ENST00000252487.5
ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr1_-_160232291 | 0.09 |
ENST00000368074.1
ENST00000447377.1 |
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr1_-_160232197 | 0.08 |
ENST00000419626.1
ENST00000610139.1 ENST00000475733.1 ENST00000407642.2 ENST00000368073.3 ENST00000326837.2 |
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr5_-_68665084 | 0.08 |
ENST00000509462.1
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr2_-_179914760 | 0.08 |
ENST00000420890.2
ENST00000409284.1 ENST00000443758.1 ENST00000446116.1 |
CCDC141
|
coiled-coil domain containing 141 |
| chr20_-_48530230 | 0.07 |
ENST00000422556.1
|
SPATA2
|
spermatogenesis associated 2 |
| chr2_+_223725652 | 0.06 |
ENST00000357430.3
ENST00000392066.3 |
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr8_+_22022653 | 0.06 |
ENST00000354870.5
ENST00000397816.3 ENST00000306349.8 |
BMP1
|
bone morphogenetic protein 1 |
| chr6_-_66417107 | 0.06 |
ENST00000370621.3
ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS
|
eyes shut homolog (Drosophila) |
| chr15_-_58358607 | 0.06 |
ENST00000249750.4
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chrX_+_49644470 | 0.05 |
ENST00000508866.2
|
USP27X
|
ubiquitin specific peptidase 27, X-linked |
| chr7_+_99717230 | 0.05 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr12_-_122879969 | 0.05 |
ENST00000540304.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr8_-_16859690 | 0.05 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr10_-_69597810 | 0.05 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr7_-_99716952 | 0.04 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr17_+_21730180 | 0.04 |
ENST00000584398.1
|
UBBP4
|
ubiquitin B pseudogene 4 |
| chr7_-_23510086 | 0.04 |
ENST00000258729.3
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr14_+_103058948 | 0.04 |
ENST00000262241.6
|
RCOR1
|
REST corepressor 1 |
| chr6_-_137314371 | 0.04 |
ENST00000432330.1
ENST00000418699.1 |
RP11-55K22.5
|
RP11-55K22.5 |
| chr20_+_52105495 | 0.04 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr14_-_94942548 | 0.03 |
ENST00000298845.7
|
SERPINA9
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 9 |
| chr20_+_31749574 | 0.03 |
ENST00000253362.2
|
BPIFA2
|
BPI fold containing family A, member 2 |
| chr15_-_88799948 | 0.03 |
ENST00000394480.2
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr1_-_227505289 | 0.03 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr6_+_33172407 | 0.03 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr5_+_140079919 | 0.03 |
ENST00000274712.3
|
ZMAT2
|
zinc finger, matrin-type 2 |
| chr6_+_123100620 | 0.02 |
ENST00000368444.3
|
FABP7
|
fatty acid binding protein 7, brain |
| chr14_-_94942527 | 0.02 |
ENST00000448305.2
|
SERPINA9
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 9 |
| chr2_-_134326009 | 0.02 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr1_-_232598163 | 0.01 |
ENST00000308942.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr17_-_42019836 | 0.01 |
ENST00000225992.3
|
PPY
|
pancreatic polypeptide |
| chr10_-_102289611 | 0.01 |
ENST00000299166.4
ENST00000370320.4 ENST00000531258.1 ENST00000370322.1 ENST00000535773.1 |
NDUFB8
SEC31B
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8, 19kDa SEC31 homolog B (S. cerevisiae) |
| chr13_-_50018241 | 0.01 |
ENST00000409308.1
|
CAB39L
|
calcium binding protein 39-like |
| chr12_-_49418407 | 0.01 |
ENST00000526209.1
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr14_-_75179774 | 0.00 |
ENST00000555249.1
ENST00000556202.1 ENST00000356357.4 ENST00000338772.5 |
AREL1
AC007956.1
|
apoptosis resistant E3 ubiquitin protein ligase 1 Full-length cDNA 5-PRIME end of clone CS0CAP004YO05 of Thymus of Homo sapiens (human); Uncharacterized protein |
| chr3_+_184056614 | 0.00 |
ENST00000453072.1
|
FAM131A
|
family with sequence similarity 131, member A |
| chr4_-_48782259 | 0.00 |
ENST00000507711.1
ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL
|
FRY-like |
| chr3_-_176914238 | 0.00 |
ENST00000430069.1
ENST00000428970.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BRCA1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.2 | 1.4 | GO:0061056 | somite specification(GO:0001757) sclerotome development(GO:0061056) |
| 0.2 | 1.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.4 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 0.7 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.1 | 0.3 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.3 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 0.4 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.3 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.1 | 1.5 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.2 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.5 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.4 | GO:0001502 | cartilage condensation(GO:0001502) high-density lipoprotein particle assembly(GO:0034380) |
| 0.0 | 0.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.7 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.2 | 0.7 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.1 | 0.4 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 1.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |