Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
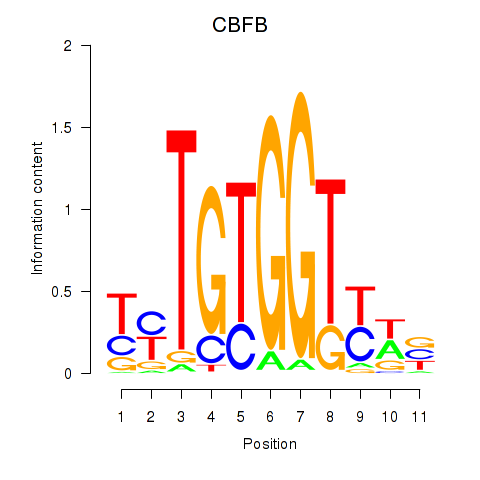
Results for CBFB
Z-value: 0.42
Transcription factors associated with CBFB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CBFB
|
ENSG00000067955.9 | core-binding factor subunit beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CBFB | hg19_v2_chr16_+_67063142_67063153 | 0.39 | 5.6e-02 | Click! |
Activity profile of CBFB motif
Sorted Z-values of CBFB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_131409476 | 2.05 |
ENST00000296871.2
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr5_+_35856951 | 1.36 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr2_+_103035102 | 1.17 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr8_+_104384616 | 1.09 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chrX_+_68048803 | 0.97 |
ENST00000204961.4
|
EFNB1
|
ephrin-B1 |
| chr21_-_36421535 | 0.82 |
ENST00000416754.1
ENST00000437180.1 ENST00000455571.1 |
RUNX1
|
runt-related transcription factor 1 |
| chr21_-_36421626 | 0.80 |
ENST00000300305.3
|
RUNX1
|
runt-related transcription factor 1 |
| chr14_-_85996332 | 0.65 |
ENST00000380722.1
|
RP11-497E19.1
|
RP11-497E19.1 |
| chr3_-_189840223 | 0.59 |
ENST00000427335.2
|
LEPREL1
|
leprecan-like 1 |
| chr6_+_29068386 | 0.54 |
ENST00000377171.3
|
OR2J1
|
olfactory receptor, family 2, subfamily J, member 1 (gene/pseudogene) |
| chr9_-_130541017 | 0.46 |
ENST00000314830.8
|
SH2D3C
|
SH2 domain containing 3C |
| chr17_+_32582293 | 0.43 |
ENST00000580907.1
ENST00000225831.4 |
CCL2
|
chemokine (C-C motif) ligand 2 |
| chr6_+_45296391 | 0.42 |
ENST00000371436.6
ENST00000576263.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr13_-_28674693 | 0.39 |
ENST00000537084.1
ENST00000241453.7 ENST00000380982.4 |
FLT3
|
fms-related tyrosine kinase 3 |
| chr1_+_89246647 | 0.34 |
ENST00000544045.1
|
PKN2
|
protein kinase N2 |
| chr12_+_15125954 | 0.30 |
ENST00000266395.2
|
PDE6H
|
phosphodiesterase 6H, cGMP-specific, cone, gamma |
| chr8_-_134072593 | 0.30 |
ENST00000427060.2
|
SLA
|
Src-like-adaptor |
| chr8_-_96281419 | 0.29 |
ENST00000286688.5
|
C8orf37
|
chromosome 8 open reading frame 37 |
| chr3_-_11610255 | 0.29 |
ENST00000424529.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr1_+_209859510 | 0.28 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr6_-_75912508 | 0.27 |
ENST00000416123.2
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr1_-_52499443 | 0.24 |
ENST00000371614.1
|
KTI12
|
KTI12 homolog, chromatin associated (S. cerevisiae) |
| chr6_+_42847649 | 0.23 |
ENST00000424341.2
ENST00000602561.1 |
RPL7L1
|
ribosomal protein L7-like 1 |
| chr9_+_15422702 | 0.21 |
ENST00000380821.3
ENST00000421710.1 |
SNAPC3
|
small nuclear RNA activating complex, polypeptide 3, 50kDa |
| chr15_+_73976545 | 0.20 |
ENST00000318443.5
ENST00000537340.2 ENST00000318424.5 |
CD276
|
CD276 molecule |
| chr2_-_105030466 | 0.19 |
ENST00000449772.1
|
AC068535.3
|
AC068535.3 |
| chr11_-_58343319 | 0.19 |
ENST00000395074.2
|
LPXN
|
leupaxin |
| chr4_+_74347400 | 0.18 |
ENST00000226355.3
|
AFM
|
afamin |
| chr2_-_64881018 | 0.18 |
ENST00000313349.3
|
SERTAD2
|
SERTA domain containing 2 |
| chr4_+_88896819 | 0.18 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr7_+_120628731 | 0.18 |
ENST00000310396.5
|
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr5_-_137878887 | 0.17 |
ENST00000507939.1
ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1
|
eukaryotic translation termination factor 1 |
| chr3_+_132843652 | 0.17 |
ENST00000508711.1
|
TMEM108
|
transmembrane protein 108 |
| chr11_-_9781068 | 0.16 |
ENST00000500698.1
|
RP11-540A21.2
|
RP11-540A21.2 |
| chr19_-_14048804 | 0.14 |
ENST00000254320.3
ENST00000586075.1 |
PODNL1
|
podocan-like 1 |
| chrX_-_49041242 | 0.14 |
ENST00000453382.1
ENST00000540849.1 ENST00000536904.1 ENST00000432913.1 |
PRICKLE3
|
prickle homolog 3 (Drosophila) |
| chr14_-_61116168 | 0.14 |
ENST00000247182.6
|
SIX1
|
SIX homeobox 1 |
| chr20_+_62492566 | 0.14 |
ENST00000369916.3
|
ABHD16B
|
abhydrolase domain containing 16B |
| chr1_-_157014865 | 0.13 |
ENST00000361409.2
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr14_-_59951112 | 0.12 |
ENST00000247194.4
|
L3HYPDH
|
L-3-hydroxyproline dehydratase (trans-) |
| chr22_+_42665742 | 0.12 |
ENST00000332965.3
ENST00000415205.1 ENST00000446578.1 |
Z83851.3
|
Z83851.3 |
| chr10_-_29811456 | 0.12 |
ENST00000535393.1
|
SVIL
|
supervillin |
| chr11_-_64885111 | 0.12 |
ENST00000528598.1
ENST00000310597.4 |
ZNHIT2
|
zinc finger, HIT-type containing 2 |
| chr19_-_14049184 | 0.12 |
ENST00000339560.5
|
PODNL1
|
podocan-like 1 |
| chr1_-_184006829 | 0.12 |
ENST00000361927.4
|
COLGALT2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr12_-_4754339 | 0.12 |
ENST00000228850.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr5_-_108745689 | 0.11 |
ENST00000361189.2
|
PJA2
|
praja ring finger 2, E3 ubiquitin protein ligase |
| chr15_+_49715293 | 0.11 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr12_-_53594227 | 0.10 |
ENST00000550743.2
|
ITGB7
|
integrin, beta 7 |
| chr6_+_27925019 | 0.10 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr11_-_66103867 | 0.09 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr4_-_156297949 | 0.08 |
ENST00000515654.1
|
MAP9
|
microtubule-associated protein 9 |
| chr3_+_157154578 | 0.08 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr14_-_59950724 | 0.08 |
ENST00000481608.1
|
L3HYPDH
|
L-3-hydroxyproline dehydratase (trans-) |
| chr19_+_10541462 | 0.07 |
ENST00000293683.5
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr4_+_71091786 | 0.07 |
ENST00000317987.5
|
FDCSP
|
follicular dendritic cell secreted protein |
| chr20_+_44637526 | 0.06 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr6_-_34524049 | 0.06 |
ENST00000374037.3
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr11_-_66103932 | 0.06 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr14_+_22748980 | 0.05 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chr2_-_207024233 | 0.04 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr6_+_52051171 | 0.04 |
ENST00000340057.1
|
IL17A
|
interleukin 17A |
| chr15_+_64386261 | 0.03 |
ENST00000560829.1
|
SNX1
|
sorting nexin 1 |
| chr8_+_67624653 | 0.03 |
ENST00000521198.2
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr21_-_36421401 | 0.02 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr6_-_34524093 | 0.02 |
ENST00000544425.1
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr11_-_66104237 | 0.01 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr1_-_240775447 | 0.00 |
ENST00000318160.4
|
GREM2
|
gremlin 2, DAN family BMP antagonist |
| chr20_+_43343886 | 0.00 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr5_+_131396222 | 0.00 |
ENST00000296870.2
|
IL3
|
interleukin 3 (colony-stimulating factor, multiple) |
| chr9_-_123342415 | 0.00 |
ENST00000349780.4
ENST00000360190.4 ENST00000360822.3 ENST00000359309.3 |
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CBFB
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.2 | 1.1 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 0.4 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 1.4 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 1.6 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.2 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.1 | 0.4 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 1.0 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.3 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.4 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.3 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.2 | GO:0016855 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.0 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 1.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |