Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
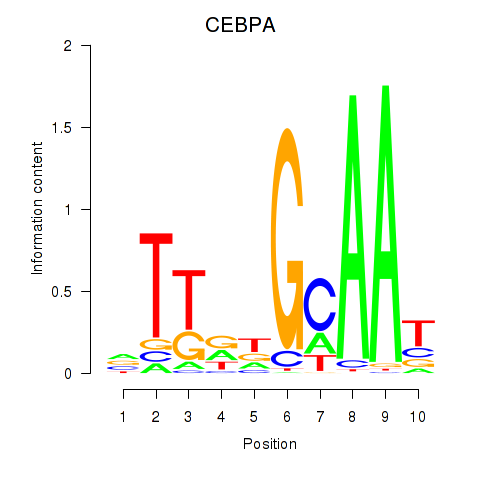
Results for CEBPA
Z-value: 0.99
Transcription factors associated with CEBPA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CEBPA
|
ENSG00000245848.2 | CCAAT enhancer binding protein alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CEBPA | hg19_v2_chr19_-_33793430_33793470 | -0.27 | 2.0e-01 | Click! |
Activity profile of CEBPA motif
Sorted Z-values of CEBPA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_102668879 | 9.57 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr6_+_127898312 | 5.97 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr1_+_101185290 | 4.88 |
ENST00000370119.4
ENST00000347652.2 ENST00000294728.2 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr1_-_173020056 | 4.33 |
ENST00000239468.2
ENST00000404377.3 |
TNFSF18
|
tumor necrosis factor (ligand) superfamily, member 18 |
| chr21_+_43619796 | 3.57 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr21_+_34775181 | 3.51 |
ENST00000290219.6
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr2_+_102721023 | 3.46 |
ENST00000409589.1
ENST00000409329.1 |
IL1R1
|
interleukin 1 receptor, type I |
| chr4_+_74606223 | 3.24 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr1_-_89531041 | 3.23 |
ENST00000370473.4
|
GBP1
|
guanylate binding protein 1, interferon-inducible |
| chr1_-_173176452 | 3.15 |
ENST00000281834.3
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr1_+_169079823 | 2.98 |
ENST00000367813.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr5_+_125758865 | 2.79 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr5_+_125758813 | 2.73 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr21_-_37852359 | 2.46 |
ENST00000399137.1
ENST00000399135.1 |
CLDN14
|
claudin 14 |
| chr18_+_61554932 | 2.40 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr5_-_121413974 | 2.40 |
ENST00000231004.4
|
LOX
|
lysyl oxidase |
| chr4_-_139163491 | 2.37 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr4_-_681114 | 2.34 |
ENST00000503156.1
|
MFSD7
|
major facilitator superfamily domain containing 7 |
| chr2_+_228678550 | 2.33 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr6_+_126240442 | 2.32 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr15_-_80263506 | 2.25 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr6_+_126102292 | 2.22 |
ENST00000368357.3
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr7_-_92777606 | 2.14 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr11_-_104905840 | 2.01 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr1_+_165797024 | 1.82 |
ENST00000372212.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr3_-_158450231 | 1.79 |
ENST00000479756.1
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr4_-_185747188 | 1.76 |
ENST00000507295.1
ENST00000504900.1 ENST00000281455.2 ENST00000454703.2 |
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr12_+_21207503 | 1.72 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr3_-_4793274 | 1.64 |
ENST00000414938.1
|
EGOT
|
eosinophil granule ontogeny transcript (non-protein coding) |
| chr3_-_107777208 | 1.60 |
ENST00000398258.3
|
CD47
|
CD47 molecule |
| chr3_-_123339343 | 1.50 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr3_-_123339418 | 1.49 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr7_+_18535786 | 1.45 |
ENST00000406072.1
|
HDAC9
|
histone deacetylase 9 |
| chr2_+_103035102 | 1.45 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chrX_-_68385354 | 1.37 |
ENST00000361478.1
|
PJA1
|
praja ring finger 1, E3 ubiquitin protein ligase |
| chrX_-_68385274 | 1.33 |
ENST00000374584.3
ENST00000590146.1 |
PJA1
|
praja ring finger 1, E3 ubiquitin protein ligase |
| chr5_-_150473127 | 1.33 |
ENST00000521001.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr6_+_31895467 | 1.31 |
ENST00000556679.1
ENST00000456570.1 |
CFB
CFB
|
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr1_-_156721502 | 1.30 |
ENST00000357325.5
|
HDGF
|
hepatoma-derived growth factor |
| chr2_-_152146385 | 1.27 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr12_+_72058130 | 1.27 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr15_+_71228826 | 1.26 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr3_+_105086056 | 1.23 |
ENST00000472644.2
|
ALCAM
|
activated leukocyte cell adhesion molecule |
| chr3_+_105085734 | 1.18 |
ENST00000306107.5
|
ALCAM
|
activated leukocyte cell adhesion molecule |
| chr1_+_165864800 | 1.13 |
ENST00000469256.2
|
UCK2
|
uridine-cytidine kinase 2 |
| chr5_-_16742330 | 1.13 |
ENST00000505695.1
ENST00000427430.2 |
MYO10
|
myosin X |
| chr3_-_112693865 | 1.11 |
ENST00000471858.1
ENST00000295863.4 ENST00000308611.3 |
CD200R1
|
CD200 receptor 1 |
| chr12_+_75874580 | 1.09 |
ENST00000456650.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr1_-_203198790 | 1.08 |
ENST00000367229.1
ENST00000255427.3 ENST00000535569.1 |
CHIT1
|
chitinase 1 (chitotriosidase) |
| chr21_+_35736302 | 1.08 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chr10_+_114133773 | 1.06 |
ENST00000354655.4
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr3_-_112218378 | 1.05 |
ENST00000334529.5
|
BTLA
|
B and T lymphocyte associated |
| chr11_-_104972158 | 1.03 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr2_+_143635067 | 1.02 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr12_+_21168630 | 1.01 |
ENST00000421593.2
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr17_+_20059302 | 0.99 |
ENST00000395530.2
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr7_+_26191809 | 0.98 |
ENST00000056233.3
|
NFE2L3
|
nuclear factor, erythroid 2-like 3 |
| chr5_+_149980622 | 0.97 |
ENST00000394243.1
|
SYNPO
|
synaptopodin |
| chr1_-_200992827 | 0.95 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr14_+_51955831 | 0.95 |
ENST00000356218.4
|
FRMD6
|
FERM domain containing 6 |
| chr4_-_69536346 | 0.94 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr19_-_56249740 | 0.93 |
ENST00000590200.1
ENST00000332836.2 |
NLRP9
|
NLR family, pyrin domain containing 9 |
| chr8_+_38585704 | 0.91 |
ENST00000519416.1
ENST00000520615.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr9_+_132934835 | 0.90 |
ENST00000372398.3
|
NCS1
|
neuronal calcium sensor 1 |
| chr3_-_112218205 | 0.89 |
ENST00000383680.4
|
BTLA
|
B and T lymphocyte associated |
| chr14_+_52164820 | 0.88 |
ENST00000554167.1
|
FRMD6
|
FERM domain containing 6 |
| chr18_-_61311485 | 0.87 |
ENST00000436264.1
ENST00000356424.6 ENST00000341074.5 |
SERPINB4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 4 |
| chr19_-_4540486 | 0.86 |
ENST00000306390.6
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr6_-_43496605 | 0.84 |
ENST00000455285.2
|
XPO5
|
exportin 5 |
| chr6_+_150920999 | 0.82 |
ENST00000367328.1
ENST00000367326.1 |
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr4_-_153274078 | 0.80 |
ENST00000263981.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr1_+_110082487 | 0.79 |
ENST00000527748.1
|
GPR61
|
G protein-coupled receptor 61 |
| chr8_+_38586068 | 0.79 |
ENST00000443286.2
ENST00000520340.1 ENST00000518415.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr7_+_30634297 | 0.79 |
ENST00000389266.3
|
GARS
|
glycyl-tRNA synthetase |
| chr16_+_56969284 | 0.79 |
ENST00000568358.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr3_-_71632894 | 0.78 |
ENST00000493089.1
|
FOXP1
|
forkhead box P1 |
| chr12_-_10251539 | 0.78 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr11_-_85397167 | 0.77 |
ENST00000316398.3
|
CCDC89
|
coiled-coil domain containing 89 |
| chr19_+_45147313 | 0.77 |
ENST00000406449.4
|
PVR
|
poliovirus receptor |
| chr6_-_11779014 | 0.75 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr15_-_89878025 | 0.75 |
ENST00000268124.5
ENST00000442287.2 |
POLG
|
polymerase (DNA directed), gamma |
| chr16_+_56485402 | 0.73 |
ENST00000566157.1
ENST00000562150.1 ENST00000561646.1 ENST00000568397.1 |
OGFOD1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr12_-_323689 | 0.71 |
ENST00000428720.1
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr17_-_67264947 | 0.71 |
ENST00000586811.1
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr12_-_10251603 | 0.70 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr17_+_20059358 | 0.70 |
ENST00000536879.1
ENST00000395522.2 ENST00000395525.3 |
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr1_+_196743912 | 0.69 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr5_-_160279207 | 0.69 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr10_-_45474237 | 0.69 |
ENST00000448778.1
ENST00000298295.3 |
C10orf10
|
chromosome 10 open reading frame 10 |
| chr3_-_66551351 | 0.68 |
ENST00000273261.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr6_+_106534192 | 0.68 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr7_+_129932974 | 0.67 |
ENST00000445470.2
ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chr1_+_159409512 | 0.67 |
ENST00000423932.3
|
OR10J1
|
olfactory receptor, family 10, subfamily J, member 1 |
| chr19_+_55014085 | 0.66 |
ENST00000351841.2
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2 |
| chr6_+_12717892 | 0.66 |
ENST00000379350.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr20_+_58630972 | 0.65 |
ENST00000313426.1
|
C20orf197
|
chromosome 20 open reading frame 197 |
| chr1_+_171154347 | 0.65 |
ENST00000209929.7
ENST00000441535.1 |
FMO2
|
flavin containing monooxygenase 2 (non-functional) |
| chr1_+_107682629 | 0.65 |
ENST00000370074.4
ENST00000370073.2 ENST00000370071.2 ENST00000542803.1 ENST00000370061.3 ENST00000370072.3 ENST00000370070.2 |
NTNG1
|
netrin G1 |
| chr8_+_145064215 | 0.65 |
ENST00000313269.5
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr12_-_10251576 | 0.63 |
ENST00000315330.4
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr19_+_55014013 | 0.63 |
ENST00000301202.2
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2 |
| chr12_+_75874460 | 0.63 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr18_-_5396271 | 0.62 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr3_-_191000172 | 0.62 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr17_+_21191341 | 0.61 |
ENST00000526076.2
ENST00000361818.5 ENST00000316920.6 |
MAP2K3
|
mitogen-activated protein kinase kinase 3 |
| chr1_+_196743943 | 0.61 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr3_-_66551397 | 0.61 |
ENST00000383703.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr15_-_70387120 | 0.60 |
ENST00000539550.1
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr11_+_86667117 | 0.60 |
ENST00000531827.1
|
RP11-736K20.6
|
RP11-736K20.6 |
| chr5_+_49962772 | 0.59 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr8_+_18248786 | 0.59 |
ENST00000520116.1
|
NAT2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr17_-_60883993 | 0.58 |
ENST00000583803.1
ENST00000456609.2 |
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr19_+_33865218 | 0.58 |
ENST00000585933.2
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr1_-_159684371 | 0.57 |
ENST00000255030.5
ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP
|
C-reactive protein, pentraxin-related |
| chr19_+_6887571 | 0.57 |
ENST00000250572.8
ENST00000381407.5 ENST00000312053.4 ENST00000450315.3 ENST00000381404.4 |
EMR1
|
egf-like module containing, mucin-like, hormone receptor-like 1 |
| chr2_-_89545079 | 0.57 |
ENST00000468494.1
|
IGKV2-30
|
immunoglobulin kappa variable 2-30 |
| chr22_+_35776828 | 0.56 |
ENST00000216117.8
|
HMOX1
|
heme oxygenase (decycling) 1 |
| chr19_-_49496557 | 0.55 |
ENST00000323798.3
ENST00000541188.1 ENST00000544287.1 ENST00000540532.1 ENST00000263276.6 |
GYS1
|
glycogen synthase 1 (muscle) |
| chr6_+_29910301 | 0.55 |
ENST00000376809.5
ENST00000376802.2 |
HLA-A
|
major histocompatibility complex, class I, A |
| chr8_+_18248755 | 0.54 |
ENST00000286479.3
|
NAT2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr9_-_117568365 | 0.54 |
ENST00000374045.4
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15 |
| chr16_+_56970567 | 0.53 |
ENST00000563911.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr16_+_23765948 | 0.53 |
ENST00000300113.2
|
CHP2
|
calcineurin-like EF-hand protein 2 |
| chr5_+_170736243 | 0.52 |
ENST00000296921.5
|
TLX3
|
T-cell leukemia homeobox 3 |
| chr1_+_248201474 | 0.52 |
ENST00000366479.2
|
OR2L2
|
olfactory receptor, family 2, subfamily L, member 2 |
| chr2_+_143635222 | 0.52 |
ENST00000375773.2
ENST00000409512.1 ENST00000410015.2 |
KYNU
|
kynureninase |
| chr16_-_2059797 | 0.52 |
ENST00000563630.1
|
ZNF598
|
zinc finger protein 598 |
| chr2_-_228497888 | 0.52 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr19_+_45394477 | 0.52 |
ENST00000252487.5
ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr10_-_27444143 | 0.51 |
ENST00000477432.1
|
YME1L1
|
YME1-like 1 ATPase |
| chr2_+_42104692 | 0.51 |
ENST00000398796.2
ENST00000442214.1 |
AC104654.1
|
AC104654.1 |
| chr11_-_119187826 | 0.51 |
ENST00000264036.4
|
MCAM
|
melanoma cell adhesion molecule |
| chr19_-_48389651 | 0.50 |
ENST00000222002.3
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr1_-_193075180 | 0.50 |
ENST00000367440.3
|
GLRX2
|
glutaredoxin 2 |
| chr5_-_145483932 | 0.50 |
ENST00000311450.4
|
PLAC8L1
|
PLAC8-like 1 |
| chr3_+_75955817 | 0.50 |
ENST00000487694.3
ENST00000602589.1 |
ROBO2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr6_+_30457244 | 0.50 |
ENST00000376630.4
|
HLA-E
|
major histocompatibility complex, class I, E |
| chrX_+_15767971 | 0.49 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chrX_+_16141667 | 0.49 |
ENST00000380289.2
|
GRPR
|
gastrin-releasing peptide receptor |
| chr17_-_60142609 | 0.49 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chr20_+_46130671 | 0.49 |
ENST00000371998.3
ENST00000371997.3 |
NCOA3
|
nuclear receptor coactivator 3 |
| chr1_-_207095324 | 0.48 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr19_-_15442701 | 0.48 |
ENST00000594841.1
ENST00000601941.1 |
BRD4
|
bromodomain containing 4 |
| chr4_+_37962018 | 0.48 |
ENST00000504686.1
|
PTTG2
|
pituitary tumor-transforming 2 |
| chr5_+_95998070 | 0.48 |
ENST00000421689.2
ENST00000510756.1 ENST00000512620.1 |
CAST
|
calpastatin |
| chr7_-_138482933 | 0.47 |
ENST00000310018.2
|
ATP6V0A4
|
ATPase, H+ transporting, lysosomal V0 subunit a4 |
| chr14_+_22554680 | 0.47 |
ENST00000390451.2
|
TRAV23DV6
|
T cell receptor alpha variable 23/delta variable 6 |
| chr17_-_6983550 | 0.47 |
ENST00000576617.1
ENST00000416562.2 |
CLEC10A
|
C-type lectin domain family 10, member A |
| chrX_+_139791917 | 0.47 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
| chr19_+_48949030 | 0.44 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr7_-_138482849 | 0.44 |
ENST00000353492.4
|
ATP6V0A4
|
ATPase, H+ transporting, lysosomal V0 subunit a4 |
| chr1_+_75594119 | 0.44 |
ENST00000294638.5
|
LHX8
|
LIM homeobox 8 |
| chr17_+_21030260 | 0.44 |
ENST00000579303.1
|
DHRS7B
|
dehydrogenase/reductase (SDR family) member 7B |
| chr7_-_14026063 | 0.43 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr5_+_156696362 | 0.43 |
ENST00000377576.3
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr17_-_6983594 | 0.43 |
ENST00000571664.1
ENST00000254868.4 |
CLEC10A
|
C-type lectin domain family 10, member A |
| chr12_-_71551652 | 0.42 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr4_+_184020398 | 0.42 |
ENST00000403733.3
ENST00000378925.3 |
WWC2
|
WW and C2 domain containing 2 |
| chr20_+_46130601 | 0.42 |
ENST00000341724.6
|
NCOA3
|
nuclear receptor coactivator 3 |
| chr2_-_239140276 | 0.42 |
ENST00000334973.4
|
AC016757.3
|
Protein LOC151174 |
| chr7_-_32111009 | 0.42 |
ENST00000396184.3
ENST00000396189.2 ENST00000321453.7 |
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr12_+_88429223 | 0.42 |
ENST00000356891.3
|
C12orf29
|
chromosome 12 open reading frame 29 |
| chr10_-_70092635 | 0.41 |
ENST00000309049.4
|
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chrX_+_153448107 | 0.41 |
ENST00000369935.5
|
OPN1MW
|
opsin 1 (cone pigments), medium-wave-sensitive |
| chr17_-_62658186 | 0.41 |
ENST00000262435.9
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr16_+_56965960 | 0.41 |
ENST00000439977.2
ENST00000344114.4 ENST00000300302.5 ENST00000379792.2 |
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr21_-_46348694 | 0.41 |
ENST00000355153.4
ENST00000397850.2 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr1_-_1711508 | 0.41 |
ENST00000378625.1
|
NADK
|
NAD kinase |
| chr12_-_53074182 | 0.40 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr11_-_10590238 | 0.40 |
ENST00000256178.3
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr5_+_95997769 | 0.40 |
ENST00000338252.3
ENST00000508830.1 |
CAST
|
calpastatin |
| chr16_+_7382745 | 0.40 |
ENST00000436368.2
ENST00000311745.5 ENST00000355637.4 ENST00000340209.4 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr1_-_153321301 | 0.39 |
ENST00000368739.3
|
PGLYRP4
|
peptidoglycan recognition protein 4 |
| chr3_-_46249878 | 0.39 |
ENST00000296140.3
|
CCR1
|
chemokine (C-C motif) receptor 1 |
| chr4_-_70826725 | 0.39 |
ENST00000353151.3
|
CSN2
|
casein beta |
| chr3_+_46395219 | 0.39 |
ENST00000445132.2
ENST00000292301.4 |
CCR2
|
chemokine (C-C motif) receptor 2 |
| chr2_-_31637560 | 0.39 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chr5_-_137878887 | 0.39 |
ENST00000507939.1
ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1
|
eukaryotic translation termination factor 1 |
| chr1_-_180991978 | 0.39 |
ENST00000542060.1
ENST00000258301.5 |
STX6
|
syntaxin 6 |
| chr10_-_61900762 | 0.39 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr2_+_90060377 | 0.38 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr10_-_79789291 | 0.38 |
ENST00000372371.3
|
POLR3A
|
polymerase (RNA) III (DNA directed) polypeptide A, 155kDa |
| chr11_-_10590118 | 0.38 |
ENST00000529598.1
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chrX_+_125953746 | 0.38 |
ENST00000371125.3
|
CXorf64
|
chromosome X open reading frame 64 |
| chr1_-_207095212 | 0.38 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr4_-_103746924 | 0.38 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr12_+_41831485 | 0.37 |
ENST00000539469.2
ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr15_-_64648273 | 0.37 |
ENST00000607537.1
ENST00000303052.7 ENST00000303032.6 |
CSNK1G1
|
casein kinase 1, gamma 1 |
| chr3_+_157154578 | 0.37 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr12_-_62586543 | 0.37 |
ENST00000416284.3
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr16_-_2059748 | 0.37 |
ENST00000562103.1
ENST00000431526.1 |
ZNF598
|
zinc finger protein 598 |
| chr14_+_95078714 | 0.37 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr10_+_114710516 | 0.36 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr11_+_60050026 | 0.36 |
ENST00000395016.3
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr7_+_117251671 | 0.36 |
ENST00000468795.1
|
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr4_-_48136217 | 0.36 |
ENST00000264316.4
|
TXK
|
TXK tyrosine kinase |
| chr1_-_8939265 | 0.36 |
ENST00000489867.1
|
ENO1
|
enolase 1, (alpha) |
| chr16_+_2587965 | 0.36 |
ENST00000342085.4
ENST00000566659.1 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr5_-_132113559 | 0.36 |
ENST00000448933.1
|
SEPT8
|
septin 8 |
| chr5_+_169010638 | 0.36 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CEBPA
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 1.1 | 4.3 | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 1.1 | 3.2 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 1.1 | 3.2 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.9 | 5.4 | GO:0032730 | positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.9 | 3.6 | GO:0055099 | detection of endogenous stimulus(GO:0009726) response to high density lipoprotein particle(GO:0055099) |
| 0.7 | 2.9 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.7 | 4.9 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.6 | 3.0 | GO:1903281 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.5 | 1.5 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.5 | 1.4 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.4 | 1.4 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.3 | 3.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 1.3 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.3 | 1.5 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.3 | 1.8 | GO:0003383 | apical constriction(GO:0003383) |
| 0.3 | 0.8 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.3 | 1.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.3 | 0.8 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.3 | 9.8 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.2 | 1.0 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.2 | 4.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 0.9 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.2 | 1.8 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.2 | 1.2 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.2 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 0.6 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.2 | 0.8 | GO:0015966 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.2 | 0.6 | GO:0014806 | heme oxidation(GO:0006788) smooth muscle hyperplasia(GO:0014806) negative regulation of mast cell cytokine production(GO:0032764) |
| 0.2 | 2.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.2 | 0.8 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.2 | 0.8 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.2 | 0.8 | GO:1901256 | interleukin-21 production(GO:0032625) macrophage colony-stimulating factor production(GO:0036301) interleukin-21 secretion(GO:0072619) regulation of macrophage colony-stimulating factor production(GO:1901256) |
| 0.1 | 1.0 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 0.7 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.1 | 0.4 | GO:0051714 | positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 0.4 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 0.4 | GO:1902567 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) negative regulation of eosinophil activation(GO:1902567) positive regulation of monocyte extravasation(GO:2000439) |
| 0.1 | 0.5 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 0.7 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 0.5 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.1 | 2.3 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 1.9 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.4 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 1.4 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.1 | 2.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 0.7 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.4 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 0.4 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.4 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 0.4 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 0.7 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.4 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 0.5 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.1 | 1.1 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 3.1 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 1.0 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 | 2.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.6 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.6 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.1 | 1.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 0.4 | GO:1904640 | response to methionine(GO:1904640) |
| 0.1 | 0.5 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.5 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.6 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.5 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.1 | 2.5 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 | 0.4 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.5 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 0.2 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.1 | 0.3 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 2.1 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.2 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 0.6 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.2 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.1 | 0.4 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.4 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.1 | 1.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.3 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 2.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.5 | GO:0042262 | DNA protection(GO:0042262) |
| 0.1 | 0.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 2.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:1901671 | positive regulation of superoxide dismutase activity(GO:1901671) positive regulation of removal of superoxide radicals(GO:1904833) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.6 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.5 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.6 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.7 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.3 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0070105 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.3 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.2 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.4 | GO:0051873 | killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.0 | 0.5 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.9 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.8 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.2 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.7 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 1.0 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.3 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.5 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.2 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.4 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.3 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.4 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.1 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 0.2 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.4 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 2.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.6 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.0 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.0 | 0.3 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.2 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.9 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.1 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.0 | 0.3 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.2 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.4 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 1.4 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.1 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.3 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.3 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.0 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.2 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.6 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.9 | GO:0050918 | positive chemotaxis(GO:0050918) |
| 0.0 | 0.1 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.2 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 1.7 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 0.1 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 1.1 | GO:0031295 | T cell costimulation(GO:0031295) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.5 | 3.0 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.3 | 1.0 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.3 | 1.7 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.3 | 0.8 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.1 | 3.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.8 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.4 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 0.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 2.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.3 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.4 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 0.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.2 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.1 | 1.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.3 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.7 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 1.4 | GO:0098553 | integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 3.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.7 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.2 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.3 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 2.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.7 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 4.6 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.2 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 1.8 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 4.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 17.4 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 7.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.9 | 3.5 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.8 | 2.3 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.7 | 3.5 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.6 | 4.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.5 | 2.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.5 | 2.4 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.3 | 3.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 1.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.3 | 0.8 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.3 | 2.9 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.3 | 1.5 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 0.6 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.2 | 0.6 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 1.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.2 | 0.5 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.2 | 1.6 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.2 | 0.5 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.2 | 3.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.6 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.1 | 2.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.4 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 2.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.6 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 0.7 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 2.6 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.7 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 2.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.4 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.8 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.8 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.4 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.1 | 0.4 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.3 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.1 | 0.3 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.3 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.1 | 0.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 1.1 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.1 | 10.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.5 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 3.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 5.6 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.1 | 0.9 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 1.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.5 | GO:0030614 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.1 | 0.2 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 0.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.6 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 1.0 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 2.0 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 4.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.5 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.4 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.6 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.4 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.8 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 1.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.2 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 1.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.4 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.0 | 0.2 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.5 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 2.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 2.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.6 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.1 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 1.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.5 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 2.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 12.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 3.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 4.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 5.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 2.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 2.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.3 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.3 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.7 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.6 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.9 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 9.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 5.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 3.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 5.0 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 3.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 11.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 2.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 3.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 0.9 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 2.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 3.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 3.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 2.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 1.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 2.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 1.7 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 2.8 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.5 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 1.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.8 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 2.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.8 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 2.4 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 1.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.6 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.5 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME DNA STRAND ELONGATION | Genes involved in DNA strand elongation |