Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
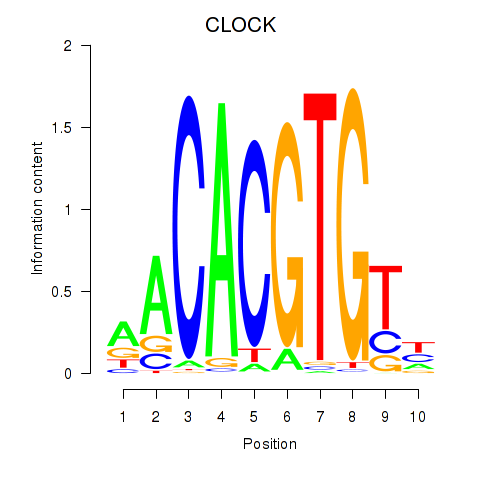
Results for CLOCK
Z-value: 0.62
Transcription factors associated with CLOCK
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CLOCK
|
ENSG00000134852.10 | clock circadian regulator |
Activity profile of CLOCK motif
Sorted Z-values of CLOCK motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_69455855 | 1.02 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr3_-_123411191 | 0.88 |
ENST00000354792.5
ENST00000508240.1 |
MYLK
|
myosin light chain kinase |
| chr6_+_7107999 | 0.75 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr3_-_127541679 | 0.66 |
ENST00000265052.5
|
MGLL
|
monoglyceride lipase |
| chr21_-_31869451 | 0.64 |
ENST00000334058.2
|
KRTAP19-4
|
keratin associated protein 19-4 |
| chrX_-_71526741 | 0.57 |
ENST00000454225.1
|
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr6_+_7107830 | 0.56 |
ENST00000379933.3
|
RREB1
|
ras responsive element binding protein 1 |
| chr6_+_7108210 | 0.52 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr9_+_706842 | 0.50 |
ENST00000382293.3
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr15_+_98503922 | 0.48 |
ENST00000268042.6
|
ARRDC4
|
arrestin domain containing 4 |
| chr19_+_46001697 | 0.48 |
ENST00000451287.2
ENST00000324688.4 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr19_+_1071203 | 0.46 |
ENST00000543365.1
|
HMHA1
|
histocompatibility (minor) HA-1 |
| chr12_+_64798095 | 0.44 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr19_+_46003056 | 0.44 |
ENST00000401593.1
ENST00000396736.2 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr3_-_127542051 | 0.42 |
ENST00000398104.1
|
MGLL
|
monoglyceride lipase |
| chr11_-_9781068 | 0.42 |
ENST00000500698.1
|
RP11-540A21.2
|
RP11-540A21.2 |
| chr4_+_95679072 | 0.42 |
ENST00000515059.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr10_-_50747064 | 0.42 |
ENST00000355832.5
ENST00000603152.1 ENST00000447839.2 |
ERCC6
PGBD3
ERCC6-PGBD3
|
excision repair cross-complementing rodent repair deficiency, complementation group 6 piggyBac transposable element derived 3 ERCC6-PGBD3 readthrough |
| chr16_+_29467127 | 0.42 |
ENST00000344620.6
|
SULT1A4
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4 |
| chr1_+_120839412 | 0.40 |
ENST00000355228.4
|
FAM72B
|
family with sequence similarity 72, member B |
| chr1_-_231376836 | 0.40 |
ENST00000451322.1
|
C1orf131
|
chromosome 1 open reading frame 131 |
| chr1_+_110163709 | 0.39 |
ENST00000369840.2
ENST00000527846.1 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr21_+_27011584 | 0.39 |
ENST00000400532.1
ENST00000480456.1 ENST00000312957.5 |
JAM2
|
junctional adhesion molecule 2 |
| chr6_+_31553978 | 0.38 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr1_-_205744205 | 0.38 |
ENST00000446390.2
|
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr5_-_102455801 | 0.38 |
ENST00000508629.1
ENST00000399004.2 |
GIN1
|
gypsy retrotransposon integrase 1 |
| chr16_+_53088885 | 0.36 |
ENST00000566029.1
ENST00000447540.1 |
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr6_+_31553901 | 0.35 |
ENST00000418507.2
ENST00000438075.2 ENST00000376100.3 ENST00000376111.4 |
LST1
|
leukocyte specific transcript 1 |
| chr2_-_231084820 | 0.35 |
ENST00000258382.5
ENST00000338556.3 |
SP110
|
SP110 nuclear body protein |
| chr2_+_201170703 | 0.35 |
ENST00000358677.5
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr1_+_152635854 | 0.35 |
ENST00000368784.1
|
LCE2D
|
late cornified envelope 2D |
| chr14_+_39736582 | 0.34 |
ENST00000556148.1
ENST00000348007.3 |
CTAGE5
|
CTAGE family, member 5 |
| chr6_+_73331918 | 0.34 |
ENST00000402622.2
ENST00000355635.3 ENST00000403813.2 ENST00000414165.2 |
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr4_-_186317034 | 0.33 |
ENST00000505916.1
|
LRP2BP
|
LRP2 binding protein |
| chr1_-_231376867 | 0.32 |
ENST00000366649.2
ENST00000318906.2 ENST00000366651.3 |
C1orf131
|
chromosome 1 open reading frame 131 |
| chr7_-_103629963 | 0.32 |
ENST00000428762.1
ENST00000343529.5 ENST00000424685.2 |
RELN
|
reelin |
| chr1_-_120935894 | 0.32 |
ENST00000369383.4
ENST00000369384.4 |
FCGR1B
|
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
| chr2_-_148778323 | 0.31 |
ENST00000440042.1
ENST00000535373.1 ENST00000540442.1 ENST00000536575.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr2_-_68384603 | 0.30 |
ENST00000406245.2
ENST00000409164.1 ENST00000295121.6 |
WDR92
|
WD repeat domain 92 |
| chr10_+_88414338 | 0.30 |
ENST00000241891.5
ENST00000443292.1 |
OPN4
|
opsin 4 |
| chr10_+_88414298 | 0.30 |
ENST00000372071.2
|
OPN4
|
opsin 4 |
| chr9_+_37120536 | 0.30 |
ENST00000336755.5
ENST00000534928.1 ENST00000322831.6 |
ZCCHC7
|
zinc finger, CCHC domain containing 7 |
| chr13_-_36920872 | 0.30 |
ENST00000451493.1
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr19_+_1067271 | 0.30 |
ENST00000536472.1
ENST00000590214.1 |
HMHA1
|
histocompatibility (minor) HA-1 |
| chr2_+_233527443 | 0.30 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr3_-_127441406 | 0.30 |
ENST00000487473.1
ENST00000484451.1 |
MGLL
|
monoglyceride lipase |
| chr7_+_1094921 | 0.30 |
ENST00000397095.1
|
GPR146
|
G protein-coupled receptor 146 |
| chr12_-_91348949 | 0.29 |
ENST00000358859.2
|
CCER1
|
coiled-coil glutamate-rich protein 1 |
| chr17_+_17942684 | 0.29 |
ENST00000376345.3
|
GID4
|
GID complex subunit 4 |
| chr17_+_17942594 | 0.29 |
ENST00000268719.4
|
GID4
|
GID complex subunit 4 |
| chrX_+_119292467 | 0.29 |
ENST00000371388.3
|
RHOXF2
|
Rhox homeobox family, member 2 |
| chr15_+_40531243 | 0.29 |
ENST00000558055.1
ENST00000455577.2 |
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr17_+_30813576 | 0.29 |
ENST00000313401.3
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr15_+_40531621 | 0.28 |
ENST00000560346.1
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr9_+_117373486 | 0.28 |
ENST00000288502.4
ENST00000374049.4 |
C9orf91
|
chromosome 9 open reading frame 91 |
| chr10_+_102756800 | 0.28 |
ENST00000370223.3
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr3_-_167813672 | 0.28 |
ENST00000470487.1
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr5_+_161274940 | 0.28 |
ENST00000393943.4
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr2_-_231084659 | 0.27 |
ENST00000258381.6
ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110
|
SP110 nuclear body protein |
| chr2_-_148778258 | 0.27 |
ENST00000392857.5
ENST00000457954.1 ENST00000392858.1 ENST00000542387.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr6_-_32811771 | 0.27 |
ENST00000395339.3
ENST00000374882.3 |
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chrX_-_119211707 | 0.27 |
ENST00000371402.2
|
RHOXF2B
|
Rhox homeobox family, member 2B |
| chr10_+_99609996 | 0.27 |
ENST00000370602.1
|
GOLGA7B
|
golgin A7 family, member B |
| chr9_-_6015607 | 0.27 |
ENST00000259569.5
|
RANBP6
|
RAN binding protein 6 |
| chr1_+_174769006 | 0.27 |
ENST00000489615.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr12_+_28343365 | 0.27 |
ENST00000545336.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr2_-_106013400 | 0.27 |
ENST00000409807.1
|
FHL2
|
four and a half LIM domains 2 |
| chr9_+_37079888 | 0.26 |
ENST00000429493.1
ENST00000593237.1 ENST00000588557.1 ENST00000430809.1 ENST00000592157.1 |
RP11-220I1.1
|
RP11-220I1.1 |
| chr1_+_62439037 | 0.26 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr11_+_61957687 | 0.26 |
ENST00000306238.3
|
SCGB1D1
|
secretoglobin, family 1D, member 1 |
| chr4_-_57301748 | 0.26 |
ENST00000264220.2
|
PPAT
|
phosphoribosyl pyrophosphate amidotransferase |
| chr3_-_98312548 | 0.26 |
ENST00000264193.2
|
CPOX
|
coproporphyrinogen oxidase |
| chr13_+_50656307 | 0.26 |
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr1_+_28655505 | 0.25 |
ENST00000373842.4
ENST00000398997.2 |
MED18
|
mediator complex subunit 18 |
| chr4_-_123542224 | 0.25 |
ENST00000264497.3
|
IL21
|
interleukin 21 |
| chr2_+_201171372 | 0.25 |
ENST00000409140.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr14_+_32030582 | 0.25 |
ENST00000550649.1
ENST00000281081.7 |
NUBPL
|
nucleotide binding protein-like |
| chr12_-_49393092 | 0.25 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr2_+_201171242 | 0.25 |
ENST00000360760.5
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr4_-_18023350 | 0.25 |
ENST00000539056.1
ENST00000382226.5 ENST00000326877.4 |
LCORL
|
ligand dependent nuclear receptor corepressor-like |
| chr17_-_73285293 | 0.24 |
ENST00000582778.1
ENST00000581988.1 ENST00000579207.1 ENST00000583332.1 ENST00000416858.2 ENST00000442286.2 ENST00000580151.1 ENST00000580994.1 ENST00000584438.1 ENST00000320362.3 ENST00000580273.1 |
SLC25A19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr2_+_201170770 | 0.24 |
ENST00000409988.3
ENST00000409385.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr18_+_72163443 | 0.24 |
ENST00000324262.4
ENST00000580672.1 |
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr13_-_36920615 | 0.24 |
ENST00000494062.2
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr17_+_55163075 | 0.24 |
ENST00000571629.1
ENST00000570423.1 ENST00000575186.1 ENST00000573085.1 ENST00000572814.1 |
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr10_+_116581503 | 0.24 |
ENST00000369248.4
ENST00000369250.3 ENST00000369246.1 |
FAM160B1
|
family with sequence similarity 160, member B1 |
| chr12_+_57623477 | 0.24 |
ENST00000557487.1
ENST00000555634.1 ENST00000556689.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr18_+_72163536 | 0.24 |
ENST00000579847.1
ENST00000583203.1 ENST00000581513.1 ENST00000577600.1 ENST00000579583.1 ENST00000584613.1 |
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr17_-_36831156 | 0.24 |
ENST00000325814.5
|
C17orf96
|
chromosome 17 open reading frame 96 |
| chr12_-_50222187 | 0.23 |
ENST00000335999.6
|
NCKAP5L
|
NCK-associated protein 5-like |
| chr1_-_11120057 | 0.23 |
ENST00000376957.2
|
SRM
|
spermidine synthase |
| chr11_+_18416103 | 0.23 |
ENST00000543445.1
ENST00000430553.2 ENST00000396222.2 ENST00000535451.1 |
LDHA
|
lactate dehydrogenase A |
| chr12_+_57623869 | 0.23 |
ENST00000414700.3
ENST00000557703.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr19_+_46806856 | 0.23 |
ENST00000300862.3
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit |
| chr22_+_22681656 | 0.23 |
ENST00000390291.2
|
IGLV1-50
|
immunoglobulin lambda variable 1-50 (non-functional) |
| chr15_+_49447947 | 0.23 |
ENST00000327171.3
ENST00000560654.1 |
GALK2
|
galactokinase 2 |
| chr1_-_3566627 | 0.23 |
ENST00000419924.2
ENST00000270708.7 |
WRAP73
|
WD repeat containing, antisense to TP73 |
| chr19_+_35168633 | 0.23 |
ENST00000505365.2
|
ZNF302
|
zinc finger protein 302 |
| chr1_-_11986442 | 0.23 |
ENST00000376572.3
ENST00000376576.3 |
KIAA2013
|
KIAA2013 |
| chr5_+_34656569 | 0.22 |
ENST00000428746.2
|
RAI14
|
retinoic acid induced 14 |
| chrX_-_71526999 | 0.22 |
ENST00000453707.2
ENST00000373619.3 |
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr9_+_96793076 | 0.22 |
ENST00000375360.3
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chr2_+_201170596 | 0.22 |
ENST00000439084.1
ENST00000409718.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr14_-_23877474 | 0.22 |
ENST00000405093.3
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr1_+_150254936 | 0.22 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr3_+_66271410 | 0.22 |
ENST00000336733.6
|
SLC25A26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr22_+_18593507 | 0.22 |
ENST00000330423.3
|
TUBA8
|
tubulin, alpha 8 |
| chr21_+_42694732 | 0.22 |
ENST00000398646.3
|
FAM3B
|
family with sequence similarity 3, member B |
| chrX_-_71526813 | 0.22 |
ENST00000246139.5
|
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr3_-_122512619 | 0.22 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr15_-_82555000 | 0.22 |
ENST00000557844.1
ENST00000359445.3 ENST00000268206.7 |
EFTUD1
|
elongation factor Tu GTP binding domain containing 1 |
| chr12_+_57624119 | 0.22 |
ENST00000555773.1
ENST00000554975.1 ENST00000449049.3 ENST00000393827.4 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr11_-_107436443 | 0.21 |
ENST00000429370.1
ENST00000417449.2 ENST00000428149.2 |
ALKBH8
|
alkB, alkylation repair homolog 8 (E. coli) |
| chr20_-_3065362 | 0.21 |
ENST00000380293.3
|
AVP
|
arginine vasopressin |
| chr17_+_72733350 | 0.21 |
ENST00000392613.5
ENST00000392612.3 ENST00000392610.1 |
RAB37
|
RAB37, member RAS oncogene family |
| chr2_-_50574856 | 0.21 |
ENST00000342183.5
|
NRXN1
|
neurexin 1 |
| chr11_+_18416133 | 0.21 |
ENST00000227157.4
ENST00000478970.2 ENST00000495052.1 |
LDHA
|
lactate dehydrogenase A |
| chr6_-_154677900 | 0.21 |
ENST00000265198.4
ENST00000520261.1 |
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chrX_-_107979616 | 0.21 |
ENST00000372129.2
|
IRS4
|
insulin receptor substrate 4 |
| chr9_+_71394945 | 0.21 |
ENST00000394264.3
|
FAM122A
|
family with sequence similarity 122A |
| chr1_-_9129895 | 0.21 |
ENST00000473209.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr19_-_18995029 | 0.20 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1 |
| chr20_-_21494654 | 0.20 |
ENST00000377142.4
|
NKX2-2
|
NK2 homeobox 2 |
| chr15_-_52944231 | 0.20 |
ENST00000546305.2
|
FAM214A
|
family with sequence similarity 214, member A |
| chr12_+_57624085 | 0.20 |
ENST00000553474.1
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr8_-_70745575 | 0.20 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr12_-_54689532 | 0.20 |
ENST00000540264.2
ENST00000312156.4 |
NFE2
|
nuclear factor, erythroid 2 |
| chr6_-_146056341 | 0.20 |
ENST00000435470.1
|
EPM2A
|
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr14_-_58894223 | 0.20 |
ENST00000555593.1
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr5_+_34656331 | 0.20 |
ENST00000265109.3
|
RAI14
|
retinoic acid induced 14 |
| chr14_-_58893832 | 0.20 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr8_-_110656995 | 0.20 |
ENST00000276646.9
ENST00000533065.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr8_-_110703819 | 0.20 |
ENST00000532779.1
ENST00000534578.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr9_-_134145880 | 0.19 |
ENST00000372269.3
ENST00000464831.1 |
FAM78A
|
family with sequence similarity 78, member A |
| chr19_-_36545649 | 0.19 |
ENST00000292894.1
|
THAP8
|
THAP domain containing 8 |
| chr2_-_231084617 | 0.19 |
ENST00000409815.2
|
SP110
|
SP110 nuclear body protein |
| chr9_+_130159504 | 0.19 |
ENST00000373352.1
ENST00000373360.3 |
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr17_-_41623716 | 0.19 |
ENST00000319349.5
|
ETV4
|
ets variant 4 |
| chr14_-_58894332 | 0.19 |
ENST00000395159.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr3_+_66271489 | 0.19 |
ENST00000536651.1
|
SLC25A26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr2_+_173420697 | 0.19 |
ENST00000282077.3
ENST00000392571.2 ENST00000410055.1 |
PDK1
|
pyruvate dehydrogenase kinase, isozyme 1 |
| chr11_+_72929402 | 0.19 |
ENST00000393596.2
|
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr8_+_109455845 | 0.19 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr8_-_102181718 | 0.19 |
ENST00000565617.1
|
KB-1460A1.5
|
KB-1460A1.5 |
| chr3_+_46283916 | 0.19 |
ENST00000395940.2
|
CCR3
|
chemokine (C-C motif) receptor 3 |
| chr5_+_161274685 | 0.19 |
ENST00000428797.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr1_-_9129598 | 0.19 |
ENST00000535586.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr1_-_205744574 | 0.18 |
ENST00000367139.3
ENST00000235932.4 ENST00000437324.2 ENST00000414729.1 |
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr9_-_86571628 | 0.18 |
ENST00000376344.3
|
C9orf64
|
chromosome 9 open reading frame 64 |
| chr6_-_146057144 | 0.18 |
ENST00000367519.3
|
EPM2A
|
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr2_+_201171064 | 0.18 |
ENST00000451764.2
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr8_-_124553437 | 0.18 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr11_+_72929319 | 0.18 |
ENST00000393597.2
ENST00000311131.2 |
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr1_+_26496362 | 0.18 |
ENST00000374266.5
ENST00000270812.5 |
ZNF593
|
zinc finger protein 593 |
| chr13_+_20207782 | 0.17 |
ENST00000414242.2
ENST00000361479.5 |
MPHOSPH8
|
M-phase phosphoprotein 8 |
| chr14_-_23876801 | 0.17 |
ENST00000356287.3
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr8_-_110704014 | 0.17 |
ENST00000529190.1
ENST00000422135.1 ENST00000419099.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr12_+_6833237 | 0.17 |
ENST00000229251.3
ENST00000539735.1 ENST00000538410.1 |
COPS7A
|
COP9 signalosome subunit 7A |
| chr1_-_45956822 | 0.17 |
ENST00000372086.3
ENST00000341771.6 |
TESK2
|
testis-specific kinase 2 |
| chr12_-_104234966 | 0.17 |
ENST00000392876.3
|
NT5DC3
|
5'-nucleotidase domain containing 3 |
| chr12_+_6833437 | 0.17 |
ENST00000534947.1
ENST00000541866.1 ENST00000534877.1 ENST00000538753.1 |
COPS7A
|
COP9 signalosome subunit 7A |
| chrX_+_23685563 | 0.17 |
ENST00000379341.4
|
PRDX4
|
peroxiredoxin 4 |
| chr14_+_78266436 | 0.17 |
ENST00000557501.1
ENST00000341211.5 |
ADCK1
|
aarF domain containing kinase 1 |
| chr7_+_105172532 | 0.17 |
ENST00000257700.2
|
RINT1
|
RAD50 interactor 1 |
| chr1_-_241520525 | 0.17 |
ENST00000366565.1
|
RGS7
|
regulator of G-protein signaling 7 |
| chr14_+_58894141 | 0.17 |
ENST00000423743.3
|
KIAA0586
|
KIAA0586 |
| chr10_-_99447024 | 0.17 |
ENST00000370626.3
|
AVPI1
|
arginine vasopressin-induced 1 |
| chr22_+_25595817 | 0.17 |
ENST00000215855.2
ENST00000404334.1 |
CRYBB3
|
crystallin, beta B3 |
| chr1_+_11333245 | 0.17 |
ENST00000376810.5
|
UBIAD1
|
UbiA prenyltransferase domain containing 1 |
| chr4_-_103266355 | 0.16 |
ENST00000424970.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr22_+_21996549 | 0.16 |
ENST00000248958.4
|
SDF2L1
|
stromal cell-derived factor 2-like 1 |
| chr14_+_58894103 | 0.16 |
ENST00000354386.6
ENST00000556134.1 |
KIAA0586
|
KIAA0586 |
| chr19_-_5340730 | 0.16 |
ENST00000372412.4
ENST00000357368.4 ENST00000262963.6 ENST00000348075.2 ENST00000353284.2 |
PTPRS
|
protein tyrosine phosphatase, receptor type, S |
| chr15_-_49103235 | 0.16 |
ENST00000380950.2
|
CEP152
|
centrosomal protein 152kDa |
| chr1_-_212873267 | 0.16 |
ENST00000243440.1
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr19_-_36545128 | 0.16 |
ENST00000538849.1
|
THAP8
|
THAP domain containing 8 |
| chr11_+_6866883 | 0.16 |
ENST00000299454.4
ENST00000379831.2 |
OR10A5
|
olfactory receptor, family 10, subfamily A, member 5 |
| chr1_-_45956800 | 0.16 |
ENST00000538496.1
|
TESK2
|
testis-specific kinase 2 |
| chr18_+_33877654 | 0.16 |
ENST00000257209.4
ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3
|
formin homology 2 domain containing 3 |
| chr9_+_130159409 | 0.16 |
ENST00000373371.3
|
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr12_+_65004292 | 0.16 |
ENST00000542104.1
ENST00000336061.2 |
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr13_+_51483814 | 0.16 |
ENST00000336617.3
ENST00000422660.1 |
RNASEH2B
|
ribonuclease H2, subunit B |
| chrX_-_128977364 | 0.16 |
ENST00000371064.3
|
ZDHHC9
|
zinc finger, DHHC-type containing 9 |
| chr20_+_42839600 | 0.16 |
ENST00000439943.1
ENST00000437730.1 |
OSER1-AS1
|
OSER1 antisense RNA 1 (head to head) |
| chr19_-_17799008 | 0.15 |
ENST00000519716.2
|
UNC13A
|
unc-13 homolog A (C. elegans) |
| chr2_+_131769256 | 0.15 |
ENST00000355771.3
|
ARHGEF4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr3_+_10857885 | 0.15 |
ENST00000254488.2
ENST00000454147.1 |
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter), member 11 |
| chr7_+_100464760 | 0.15 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr1_+_156830678 | 0.15 |
ENST00000524377.1
ENST00000358660.3 |
NTRK1
|
neurotrophic tyrosine kinase, receptor, type 1 |
| chr11_+_45792967 | 0.15 |
ENST00000378779.2
|
CTD-2210P24.4
|
Uncharacterized protein |
| chr20_+_24929866 | 0.15 |
ENST00000480798.1
ENST00000376835.2 |
CST7
|
cystatin F (leukocystatin) |
| chr14_-_47120956 | 0.15 |
ENST00000298283.3
|
RPL10L
|
ribosomal protein L10-like |
| chr1_-_67142710 | 0.15 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr1_+_156830607 | 0.15 |
ENST00000368196.3
|
NTRK1
|
neurotrophic tyrosine kinase, receptor, type 1 |
| chr20_+_42839722 | 0.15 |
ENST00000442383.1
ENST00000435163.1 |
OSER1-AS1
|
OSER1 antisense RNA 1 (head to head) |
| chr6_-_34113856 | 0.15 |
ENST00000538487.2
|
GRM4
|
glutamate receptor, metabotropic 4 |
| chrX_-_14047996 | 0.14 |
ENST00000380523.4
ENST00000398355.3 |
GEMIN8
|
gem (nuclear organelle) associated protein 8 |
| chr9_+_124088860 | 0.14 |
ENST00000373806.1
|
GSN
|
gelsolin |
| chrX_+_118108601 | 0.14 |
ENST00000371628.3
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr5_+_140588269 | 0.14 |
ENST00000541609.1
ENST00000239450.2 |
PCDHB12
|
protocadherin beta 12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CLOCK
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.8 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.2 | 1.4 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 1.0 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.2 | 0.5 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.5 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 0.9 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 1.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.4 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.9 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.3 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.3 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.4 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.2 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.1 | 0.2 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.2 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 0.4 | GO:0098704 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.1 | 0.3 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.1 | 0.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.2 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.1 | 0.6 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.3 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.2 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.1 | 0.3 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.4 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.4 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.0 | 0.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.3 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0032759 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.0 | 0.1 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.4 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.0 | 0.3 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.2 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.2 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.0 | 0.1 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.4 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.4 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.0 | 0.3 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.2 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.2 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0060849 | radial pattern formation(GO:0009956) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 0.4 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.0 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.3 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:1904450 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) positive regulation of prolactin secretion(GO:1902722) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.3 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.2 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.0 | GO:0003250 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) |
| 0.0 | 0.2 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0072299 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.0 | 0.1 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.0 | GO:1904020 | positive regulation of adenosine receptor signaling pathway(GO:0060168) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.0 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.0 | 0.3 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.1 | GO:0042723 | thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.0 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.1 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.0 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.1 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.2 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.1 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.9 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.3 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.1 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.6 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.6 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.6 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.7 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.0 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 2.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.0 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.1 | 0.9 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 0.3 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.1 | 0.3 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 0.6 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.3 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 0.2 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.1 | 1.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.2 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 0.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.4 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 1.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.4 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.1 | 0.5 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.2 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.1 | 0.4 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.1 | 0.3 | GO:0010465 | neurotrophin p75 receptor binding(GO:0005166) nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.2 | GO:0031177 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.2 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.2 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.3 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.1 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 1.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.5 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.3 | GO:0005536 | glucose binding(GO:0005536) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 1.1 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.0 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.0 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.0 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.0 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 1.0 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |