Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
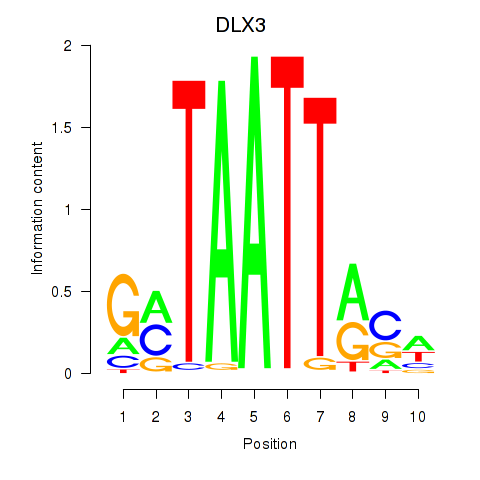
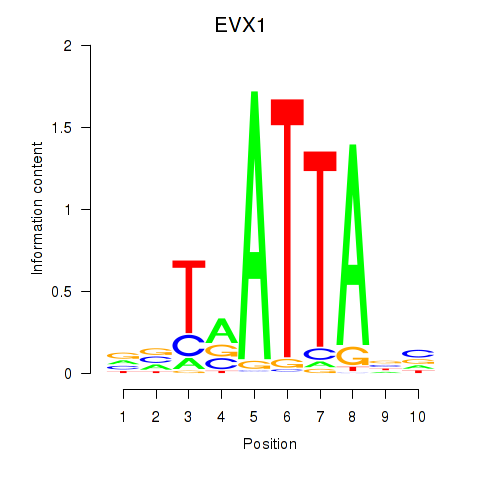
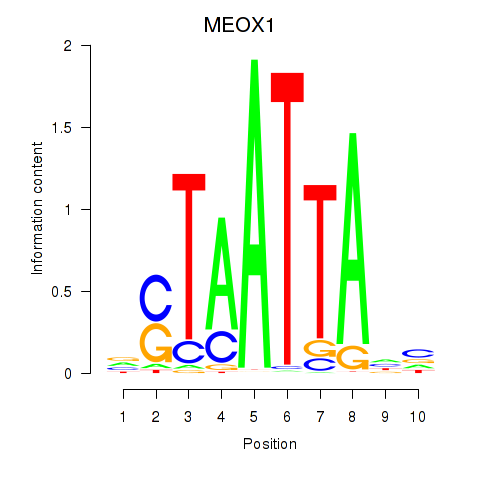
Results for DLX3_EVX1_MEOX1
Z-value: 0.39
Transcription factors associated with DLX3_EVX1_MEOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DLX3
|
ENSG00000064195.7 | distal-less homeobox 3 |
|
EVX1
|
ENSG00000106038.8 | even-skipped homeobox 1 |
|
MEOX1
|
ENSG00000005102.8 | mesenchyme homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EVX1 | hg19_v2_chr7_+_27282319_27282394 | -0.39 | 5.5e-02 | Click! |
| DLX3 | hg19_v2_chr17_-_48072574_48072597 | -0.24 | 2.6e-01 | Click! |
| MEOX1 | hg19_v2_chr17_-_41738931_41739040, hg19_v2_chr17_-_41739283_41739322 | 0.22 | 3.0e-01 | Click! |
Activity profile of DLX3_EVX1_MEOX1 motif
Sorted Z-values of DLX3_EVX1_MEOX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_141747950 | 0.61 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr5_-_141249154 | 0.42 |
ENST00000357517.5
ENST00000536585.1 |
PCDH1
|
protocadherin 1 |
| chr3_+_2933893 | 0.41 |
ENST00000397459.2
|
CNTN4
|
contactin 4 |
| chr6_+_29274403 | 0.40 |
ENST00000377160.2
|
OR14J1
|
olfactory receptor, family 14, subfamily J, member 1 |
| chr15_-_55562582 | 0.40 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr1_+_101003687 | 0.34 |
ENST00000315033.4
|
GPR88
|
G protein-coupled receptor 88 |
| chr12_-_50643664 | 0.33 |
ENST00000550592.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr18_-_33709268 | 0.30 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr15_-_55562479 | 0.30 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr15_-_55563072 | 0.30 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr5_+_66300446 | 0.28 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr10_+_118083919 | 0.27 |
ENST00000333254.3
|
CCDC172
|
coiled-coil domain containing 172 |
| chrX_-_21676442 | 0.26 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr21_-_32253874 | 0.26 |
ENST00000332378.4
|
KRTAP11-1
|
keratin associated protein 11-1 |
| chr17_+_43238438 | 0.26 |
ENST00000593138.1
ENST00000586681.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr15_+_89631647 | 0.25 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr13_-_46716969 | 0.23 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr15_+_41913690 | 0.22 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr4_+_144354644 | 0.21 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr20_+_60174827 | 0.20 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr7_+_99717230 | 0.20 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr2_+_196313239 | 0.19 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr19_-_7968427 | 0.19 |
ENST00000539278.1
|
AC010336.1
|
Uncharacterized protein |
| chr3_-_151034734 | 0.19 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr4_+_71458012 | 0.18 |
ENST00000449493.2
|
AMBN
|
ameloblastin (enamel matrix protein) |
| chr2_+_169757750 | 0.18 |
ENST00000375363.3
ENST00000429379.2 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase, catalytic, 2 |
| chr3_+_111717511 | 0.18 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr7_-_92777606 | 0.18 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr12_-_71182695 | 0.18 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr5_-_159827033 | 0.18 |
ENST00000523213.1
|
C5orf54
|
chromosome 5 open reading frame 54 |
| chr3_+_111717600 | 0.17 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr20_-_50418972 | 0.17 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr6_-_87804815 | 0.16 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr20_-_50418947 | 0.16 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr7_-_81399411 | 0.16 |
ENST00000423064.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr4_-_119759795 | 0.15 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr3_+_111718036 | 0.15 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr6_-_76203454 | 0.15 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr4_+_66536248 | 0.15 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr16_+_53133070 | 0.15 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr18_+_55888767 | 0.14 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr19_-_47137942 | 0.14 |
ENST00000300873.4
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr12_-_86650045 | 0.14 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr12_-_6233828 | 0.14 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr15_-_37393406 | 0.14 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr20_-_50419055 | 0.14 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr15_+_75080883 | 0.13 |
ENST00000567571.1
|
CSK
|
c-src tyrosine kinase |
| chr12_+_81110684 | 0.13 |
ENST00000228644.3
|
MYF5
|
myogenic factor 5 |
| chr4_+_88754069 | 0.13 |
ENST00000395102.4
ENST00000497649.2 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr4_+_88754113 | 0.13 |
ENST00000560249.1
ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr7_-_92855762 | 0.12 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr4_+_71457970 | 0.12 |
ENST00000322937.6
|
AMBN
|
ameloblastin (enamel matrix protein) |
| chr4_-_139163491 | 0.12 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr4_-_66536057 | 0.11 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr14_+_22748980 | 0.11 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chr22_-_41682172 | 0.11 |
ENST00000356244.3
|
RANGAP1
|
Ran GTPase activating protein 1 |
| chr11_-_26593779 | 0.11 |
ENST00000529533.1
|
MUC15
|
mucin 15, cell surface associated |
| chr12_-_53045948 | 0.11 |
ENST00000309680.3
|
KRT2
|
keratin 2 |
| chr15_-_100882191 | 0.11 |
ENST00000268070.4
|
ADAMTS17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr7_-_25019760 | 0.11 |
ENST00000352860.1
ENST00000353930.1 ENST00000431825.2 ENST00000313367.2 |
OSBPL3
|
oxysterol binding protein-like 3 |
| chr11_-_26593677 | 0.10 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr4_-_174451370 | 0.10 |
ENST00000359562.4
|
HAND2
|
heart and neural crest derivatives expressed 2 |
| chr14_-_51027838 | 0.10 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr6_-_76203345 | 0.10 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr7_+_154002189 | 0.09 |
ENST00000332007.3
|
DPP6
|
dipeptidyl-peptidase 6 |
| chr6_-_55739542 | 0.09 |
ENST00000446683.2
|
BMP5
|
bone morphogenetic protein 5 |
| chr20_+_43990576 | 0.09 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr6_+_155537771 | 0.09 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr6_-_36515177 | 0.09 |
ENST00000229812.7
|
STK38
|
serine/threonine kinase 38 |
| chr17_-_18430160 | 0.09 |
ENST00000392176.3
|
FAM106A
|
family with sequence similarity 106, member A |
| chr1_-_67266939 | 0.08 |
ENST00000304526.2
|
INSL5
|
insulin-like 5 |
| chr11_-_70672645 | 0.08 |
ENST00000423696.2
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr8_+_22424551 | 0.08 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr6_+_127898312 | 0.08 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr14_+_64680854 | 0.08 |
ENST00000458046.2
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr2_-_61697862 | 0.08 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr15_-_31393910 | 0.08 |
ENST00000397795.2
ENST00000256552.6 ENST00000559179.1 |
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr3_-_33686925 | 0.08 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr17_-_39165366 | 0.08 |
ENST00000391588.1
|
KRTAP3-1
|
keratin associated protein 3-1 |
| chr14_+_56584414 | 0.07 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr8_-_82395461 | 0.07 |
ENST00000256104.4
|
FABP4
|
fatty acid binding protein 4, adipocyte |
| chrX_-_118986911 | 0.07 |
ENST00000276201.2
ENST00000345865.2 |
UPF3B
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr6_-_31107127 | 0.07 |
ENST00000259845.4
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chr9_+_82188077 | 0.07 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr5_-_95297678 | 0.06 |
ENST00000237853.4
|
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr5_-_34043310 | 0.06 |
ENST00000231338.7
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3 |
| chr2_-_203735586 | 0.06 |
ENST00000454326.1
ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr5_-_95297534 | 0.06 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr17_-_39203519 | 0.06 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr4_-_87028478 | 0.06 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr8_+_39792474 | 0.06 |
ENST00000502986.2
|
IDO2
|
indoleamine 2,3-dioxygenase 2 |
| chr14_+_22984601 | 0.06 |
ENST00000390509.1
|
TRAJ28
|
T cell receptor alpha joining 28 |
| chr6_-_116833500 | 0.06 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr13_+_36050881 | 0.06 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr2_+_113763031 | 0.06 |
ENST00000259211.6
|
IL36A
|
interleukin 36, alpha |
| chr4_+_41614720 | 0.06 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr3_+_185431080 | 0.06 |
ENST00000296270.1
|
C3orf65
|
chromosome 3 open reading frame 65 |
| chr4_+_41614909 | 0.06 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr8_+_38831683 | 0.06 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chr22_-_32860427 | 0.06 |
ENST00000534972.1
ENST00000397450.1 ENST00000397452.1 |
BPIFC
|
BPI fold containing family C |
| chrX_+_78426469 | 0.05 |
ENST00000276077.1
|
GPR174
|
G protein-coupled receptor 174 |
| chr4_-_46126093 | 0.05 |
ENST00000295452.4
|
GABRG1
|
gamma-aminobutyric acid (GABA) A receptor, gamma 1 |
| chr9_+_125132803 | 0.05 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr4_-_105416039 | 0.05 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr15_-_34875771 | 0.05 |
ENST00000267731.7
|
GOLGA8B
|
golgin A8 family, member B |
| chr8_+_7783859 | 0.05 |
ENST00000400120.3
|
ZNF705B
|
zinc finger protein 705B |
| chr15_+_63188009 | 0.05 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr8_+_92261516 | 0.05 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr10_+_114710211 | 0.05 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr18_-_71959159 | 0.05 |
ENST00000494131.2
ENST00000397914.4 ENST00000340533.4 |
CYB5A
|
cytochrome b5 type A (microsomal) |
| chr10_+_118187379 | 0.05 |
ENST00000369230.3
|
PNLIPRP3
|
pancreatic lipase-related protein 3 |
| chr1_+_68150744 | 0.05 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr12_+_15699286 | 0.05 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr2_-_207082748 | 0.05 |
ENST00000407325.2
ENST00000411719.1 |
GPR1
|
G protein-coupled receptor 1 |
| chr10_+_24738355 | 0.05 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr2_-_55647057 | 0.05 |
ENST00000436346.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr6_-_76072719 | 0.05 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr8_+_70404996 | 0.05 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr1_+_236958554 | 0.05 |
ENST00000366577.5
ENST00000418145.2 |
MTR
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr2_-_55646957 | 0.05 |
ENST00000263630.8
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr1_-_68698197 | 0.05 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr3_+_44840679 | 0.05 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chr7_-_81399438 | 0.05 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr14_+_100485712 | 0.05 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr3_-_47934234 | 0.04 |
ENST00000420772.2
|
MAP4
|
microtubule-associated protein 4 |
| chr6_+_39760129 | 0.04 |
ENST00000274867.4
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr1_-_28384598 | 0.04 |
ENST00000373864.1
|
EYA3
|
eyes absent homolog 3 (Drosophila) |
| chr11_-_33913708 | 0.04 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr4_-_120243545 | 0.04 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr15_+_93443419 | 0.04 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr2_-_17981462 | 0.04 |
ENST00000402989.1
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr11_-_36619771 | 0.04 |
ENST00000311485.3
ENST00000527033.1 ENST00000532616.1 |
RAG2
|
recombination activating gene 2 |
| chr4_-_39979576 | 0.04 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr5_-_142780280 | 0.04 |
ENST00000424646.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr5_+_140625147 | 0.04 |
ENST00000231173.3
|
PCDHB15
|
protocadherin beta 15 |
| chr4_-_109684120 | 0.04 |
ENST00000512646.1
ENST00000411864.2 ENST00000296486.3 ENST00000510706.1 |
ETNPPL
|
ethanolamine-phosphate phospho-lyase |
| chr6_-_55740352 | 0.04 |
ENST00000370830.3
|
BMP5
|
bone morphogenetic protein 5 |
| chr11_-_117747327 | 0.04 |
ENST00000584230.1
ENST00000527429.1 ENST00000584394.1 ENST00000532984.1 |
FXYD6
FXYD6-FXYD2
|
FXYD domain containing ion transport regulator 6 FXYD6-FXYD2 readthrough |
| chr4_-_66536196 | 0.04 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr11_+_128563652 | 0.04 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr2_-_190927447 | 0.04 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr9_-_13165457 | 0.04 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr16_-_20338748 | 0.04 |
ENST00000575582.1
ENST00000341642.5 ENST00000381362.4 ENST00000572347.1 ENST00000572478.1 ENST00000302555.5 |
GP2
|
glycoprotein 2 (zymogen granule membrane) |
| chr6_+_50061315 | 0.04 |
ENST00000415106.1
|
RP11-397G17.1
|
RP11-397G17.1 |
| chr17_-_39306054 | 0.03 |
ENST00000343246.4
|
KRTAP4-5
|
keratin associated protein 4-5 |
| chr1_-_94586651 | 0.03 |
ENST00000535735.1
ENST00000370225.3 |
ABCA4
|
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr21_-_43346790 | 0.03 |
ENST00000329623.7
|
C2CD2
|
C2 calcium-dependent domain containing 2 |
| chr16_-_67517716 | 0.03 |
ENST00000290953.2
|
AGRP
|
agouti related protein homolog (mouse) |
| chr17_+_62223320 | 0.03 |
ENST00000580828.1
ENST00000582965.1 |
SNORA76
|
small nucleolar RNA, H/ACA box 76 |
| chr7_-_33102338 | 0.03 |
ENST00000610140.1
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr13_-_95131923 | 0.03 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr7_-_33102399 | 0.03 |
ENST00000242210.7
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr14_-_69261310 | 0.03 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr4_-_74486109 | 0.03 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr3_+_4345043 | 0.03 |
ENST00000430981.1
ENST00000425863.1 |
SETMAR
|
SET domain and mariner transposase fusion gene |
| chrX_+_135730297 | 0.03 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr11_+_101918153 | 0.03 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr9_+_124329336 | 0.03 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr6_+_167536230 | 0.03 |
ENST00000341935.5
ENST00000349984.4 |
CCR6
|
chemokine (C-C motif) receptor 6 |
| chr7_-_99381798 | 0.03 |
ENST00000415003.1
ENST00000354593.2 |
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4 |
| chrX_-_19988382 | 0.03 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chr17_+_60536002 | 0.03 |
ENST00000582809.1
|
TLK2
|
tousled-like kinase 2 |
| chr7_+_50348268 | 0.03 |
ENST00000438033.1
ENST00000439701.1 |
IKZF1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr15_+_89631381 | 0.03 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chr3_+_4344988 | 0.03 |
ENST00000358065.4
|
SETMAR
|
SET domain and mariner transposase fusion gene |
| chr4_-_74486347 | 0.03 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr20_-_49307897 | 0.03 |
ENST00000535356.1
|
FAM65C
|
family with sequence similarity 65, member C |
| chr6_+_47666275 | 0.03 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr17_-_39150385 | 0.03 |
ENST00000391586.1
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr15_+_64680003 | 0.03 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr4_-_25865159 | 0.03 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr11_+_67219867 | 0.03 |
ENST00000438189.2
|
CABP4
|
calcium binding protein 4 |
| chr1_-_50889155 | 0.03 |
ENST00000404795.3
|
DMRTA2
|
DMRT-like family A2 |
| chr11_-_107729887 | 0.03 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr15_+_89164520 | 0.03 |
ENST00000332810.3
|
AEN
|
apoptosis enhancing nuclease |
| chr2_+_171034646 | 0.03 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr10_-_23633720 | 0.03 |
ENST00000323327.4
|
C10orf67
|
chromosome 10 open reading frame 67 |
| chr13_-_30881134 | 0.03 |
ENST00000380617.3
ENST00000441394.1 |
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr10_-_69455873 | 0.03 |
ENST00000433211.2
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3 |
| chr20_+_44258165 | 0.03 |
ENST00000372643.3
|
WFDC10A
|
WAP four-disulfide core domain 10A |
| chr6_-_72130472 | 0.03 |
ENST00000426635.2
|
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr10_-_33623310 | 0.03 |
ENST00000395995.1
ENST00000374823.5 ENST00000374821.5 ENST00000374816.3 |
NRP1
|
neuropilin 1 |
| chr19_+_50016610 | 0.02 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr1_+_152943122 | 0.02 |
ENST00000328051.2
|
SPRR4
|
small proline-rich protein 4 |
| chr7_+_75931861 | 0.02 |
ENST00000248553.6
|
HSPB1
|
heat shock 27kDa protein 1 |
| chr12_-_78753496 | 0.02 |
ENST00000548512.1
|
RP11-38F22.1
|
RP11-38F22.1 |
| chr8_-_7243080 | 0.02 |
ENST00000400156.4
|
ZNF705G
|
zinc finger protein 705G |
| chr17_-_27418537 | 0.02 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr14_+_97925151 | 0.02 |
ENST00000554862.1
ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1
|
CTD-2506J14.1 |
| chr3_-_195538728 | 0.02 |
ENST00000349607.4
ENST00000346145.4 |
MUC4
|
mucin 4, cell surface associated |
| chr7_-_151217166 | 0.02 |
ENST00000496004.1
|
RHEB
|
Ras homolog enriched in brain |
| chr2_-_68052694 | 0.02 |
ENST00000457448.1
|
AC010987.6
|
AC010987.6 |
| chr2_-_31361543 | 0.02 |
ENST00000349752.5
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr3_+_159557637 | 0.02 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr2_-_216003127 | 0.02 |
ENST00000412081.1
ENST00000272895.7 |
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr13_+_73632897 | 0.02 |
ENST00000377687.4
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr3_+_35722424 | 0.02 |
ENST00000396481.2
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr1_+_62439037 | 0.02 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr6_-_27880174 | 0.02 |
ENST00000303324.2
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2 |
| chr11_-_117748138 | 0.02 |
ENST00000527717.1
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DLX3_EVX1_MEOX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.0 | 0.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) visceral serous pericardium development(GO:0061032) |
| 0.0 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.0 | GO:0090260 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.0 | 0.1 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.0 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.0 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.0 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.0 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.0 | GO:1990032 | parallel fiber(GO:1990032) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.3 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 1.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.0 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |