Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
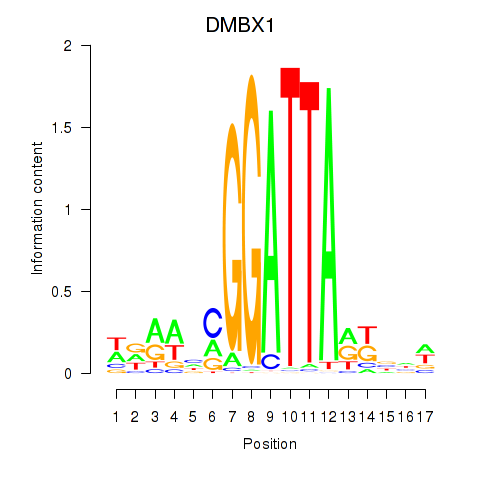
Results for DMBX1
Z-value: 0.37
Transcription factors associated with DMBX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DMBX1
|
ENSG00000197587.6 | diencephalon/mesencephalon homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DMBX1 | hg19_v2_chr1_+_46972668_46972669 | -0.17 | 4.3e-01 | Click! |
Activity profile of DMBX1 motif
Sorted Z-values of DMBX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_137851220 | 0.38 |
ENST00000236709.3
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr2_-_165812028 | 0.26 |
ENST00000303735.4
|
SLC38A11
|
solute carrier family 38, member 11 |
| chr2_-_165811983 | 0.26 |
ENST00000409058.1
ENST00000409149.3 |
SLC38A11
|
solute carrier family 38, member 11 |
| chr19_-_6459746 | 0.26 |
ENST00000301454.4
ENST00000334510.5 |
SLC25A23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23 |
| chr12_-_10959892 | 0.23 |
ENST00000240615.2
|
TAS2R8
|
taste receptor, type 2, member 8 |
| chrX_-_102565932 | 0.21 |
ENST00000372674.1
ENST00000372677.3 |
BEX2
|
brain expressed X-linked 2 |
| chr2_-_70780572 | 0.18 |
ENST00000450929.1
|
TGFA
|
transforming growth factor, alpha |
| chr12_-_56727676 | 0.18 |
ENST00000547572.1
ENST00000257931.5 ENST00000440411.3 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr2_-_165811756 | 0.18 |
ENST00000409662.1
|
SLC38A11
|
solute carrier family 38, member 11 |
| chr12_+_53693812 | 0.18 |
ENST00000549488.1
|
C12orf10
|
chromosome 12 open reading frame 10 |
| chrX_-_32173579 | 0.17 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr12_-_56727487 | 0.17 |
ENST00000548043.1
ENST00000425394.2 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr20_+_59654146 | 0.17 |
ENST00000441660.1
|
RP5-827L5.1
|
RP5-827L5.1 |
| chr3_+_188664988 | 0.16 |
ENST00000433971.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr3_-_49058479 | 0.16 |
ENST00000440857.1
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr1_-_13452656 | 0.16 |
ENST00000376132.3
|
PRAMEF13
|
PRAME family member 13 |
| chr1_+_162351503 | 0.15 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr4_+_619347 | 0.15 |
ENST00000255622.6
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr12_-_118628350 | 0.15 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr6_+_130339710 | 0.13 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr11_+_61248583 | 0.13 |
ENST00000432063.2
ENST00000338608.2 |
PPP1R32
|
protein phosphatase 1, regulatory subunit 32 |
| chr4_-_54232144 | 0.13 |
ENST00000388940.4
ENST00000503450.1 ENST00000401642.3 |
SCFD2
|
sec1 family domain containing 2 |
| chr12_-_56321397 | 0.13 |
ENST00000557259.1
ENST00000549939.1 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr7_+_21582638 | 0.13 |
ENST00000409508.3
ENST00000328843.6 |
DNAH11
|
dynein, axonemal, heavy chain 11 |
| chr1_-_94586651 | 0.13 |
ENST00000535735.1
ENST00000370225.3 |
ABCA4
|
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr13_-_62001982 | 0.13 |
ENST00000409186.1
|
PCDH20
|
protocadherin 20 |
| chr4_-_170679024 | 0.12 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27 |
| chr3_-_121468513 | 0.12 |
ENST00000494517.1
ENST00000393667.3 |
GOLGB1
|
golgin B1 |
| chr5_+_175740083 | 0.12 |
ENST00000332772.4
|
SIMC1
|
SUMO-interacting motifs containing 1 |
| chr10_-_101491828 | 0.12 |
ENST00000370483.5
ENST00000016171.5 |
COX15
|
cytochrome c oxidase assembly homolog 15 (yeast) |
| chr7_+_1727755 | 0.12 |
ENST00000424383.2
|
ELFN1
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1 |
| chr3_+_138340049 | 0.12 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr3_-_121468602 | 0.11 |
ENST00000340645.5
|
GOLGB1
|
golgin B1 |
| chr10_+_103986085 | 0.11 |
ENST00000370005.3
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chr3_+_138340067 | 0.10 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr14_+_50234309 | 0.10 |
ENST00000298307.5
|
KLHDC2
|
kelch domain containing 2 |
| chr2_-_14541060 | 0.10 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr7_-_135412925 | 0.09 |
ENST00000354042.4
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr19_+_41284121 | 0.09 |
ENST00000594800.1
ENST00000357052.2 ENST00000602173.1 |
RAB4B
|
RAB4B, member RAS oncogene family |
| chr12_+_115800817 | 0.09 |
ENST00000547948.1
|
RP11-116D17.1
|
HCG2038717; Uncharacterized protein |
| chr6_-_111927062 | 0.09 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr17_+_4643337 | 0.09 |
ENST00000592813.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr3_-_196242233 | 0.08 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr16_-_31076273 | 0.08 |
ENST00000426488.2
|
ZNF668
|
zinc finger protein 668 |
| chr1_+_120839005 | 0.08 |
ENST00000369390.3
ENST00000452190.1 |
FAM72B
|
family with sequence similarity 72, member B |
| chr17_+_4643300 | 0.08 |
ENST00000433935.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr17_-_19651654 | 0.08 |
ENST00000395555.3
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr12_-_56321649 | 0.08 |
ENST00000454792.2
ENST00000408946.2 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr11_-_45940343 | 0.08 |
ENST00000532681.1
|
PEX16
|
peroxisomal biogenesis factor 16 |
| chr17_-_19651668 | 0.07 |
ENST00000494157.2
ENST00000225740.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr1_+_206138457 | 0.07 |
ENST00000367128.3
ENST00000431655.2 |
FAM72A
|
family with sequence similarity 72, member A |
| chr6_-_26250835 | 0.07 |
ENST00000446824.2
|
HIST1H3F
|
histone cluster 1, H3f |
| chr4_+_140586922 | 0.07 |
ENST00000265498.1
ENST00000506797.1 |
MGST2
|
microsomal glutathione S-transferase 2 |
| chr16_-_31076332 | 0.07 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr1_-_143913143 | 0.06 |
ENST00000400889.1
|
FAM72D
|
family with sequence similarity 72, member D |
| chr19_-_17186229 | 0.06 |
ENST00000253669.5
ENST00000448593.2 |
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr3_-_149470229 | 0.06 |
ENST00000473414.1
|
COMMD2
|
COMM domain containing 2 |
| chr10_-_35104185 | 0.06 |
ENST00000374789.3
ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3
|
par-3 family cell polarity regulator |
| chr4_+_619386 | 0.06 |
ENST00000496514.1
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr19_-_17185848 | 0.06 |
ENST00000593360.1
|
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr17_-_4643114 | 0.05 |
ENST00000293778.6
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chrX_-_71792477 | 0.05 |
ENST00000421523.1
ENST00000415409.1 ENST00000373559.4 ENST00000373556.3 ENST00000373560.2 ENST00000373583.1 ENST00000429103.2 ENST00000373571.1 ENST00000373554.1 |
HDAC8
|
histone deacetylase 8 |
| chr3_-_19975665 | 0.05 |
ENST00000295824.9
ENST00000389256.4 |
EFHB
|
EF-hand domain family, member B |
| chr17_+_68100989 | 0.04 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr20_+_34556488 | 0.04 |
ENST00000373973.3
ENST00000349339.1 ENST00000538900.1 |
CNBD2
|
cyclic nucleotide binding domain containing 2 |
| chr17_-_48207157 | 0.04 |
ENST00000330175.4
ENST00000503131.1 |
SAMD14
|
sterile alpha motif domain containing 14 |
| chr19_-_51538118 | 0.04 |
ENST00000529888.1
|
KLK12
|
kallikrein-related peptidase 12 |
| chr2_+_37571845 | 0.04 |
ENST00000537448.1
|
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr3_-_16524357 | 0.04 |
ENST00000432519.1
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr15_-_61521495 | 0.03 |
ENST00000335670.6
|
RORA
|
RAR-related orphan receptor A |
| chr19_-_51538148 | 0.03 |
ENST00000319590.4
ENST00000250351.4 |
KLK12
|
kallikrein-related peptidase 12 |
| chr1_-_16563641 | 0.03 |
ENST00000375599.3
|
RSG1
|
REM2 and RAB-like small GTPase 1 |
| chr17_+_60536002 | 0.03 |
ENST00000582809.1
|
TLK2
|
tousled-like kinase 2 |
| chr6_-_138893661 | 0.03 |
ENST00000427025.2
|
NHSL1
|
NHS-like 1 |
| chrX_-_48931648 | 0.03 |
ENST00000376386.3
ENST00000376390.4 |
PRAF2
|
PRA1 domain family, member 2 |
| chr21_-_35016231 | 0.02 |
ENST00000438788.1
|
CRYZL1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr6_+_89791507 | 0.02 |
ENST00000354922.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr1_+_12851545 | 0.02 |
ENST00000332296.7
|
PRAMEF1
|
PRAME family member 1 |
| chr17_+_48911942 | 0.02 |
ENST00000426127.1
|
WFIKKN2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr19_+_17448348 | 0.02 |
ENST00000324894.8
ENST00000358792.7 ENST00000600625.1 |
GTPBP3
|
GTP binding protein 3 (mitochondrial) |
| chr12_-_103310987 | 0.02 |
ENST00000307000.2
|
PAH
|
phenylalanine hydroxylase |
| chr2_+_73461410 | 0.02 |
ENST00000399032.2
ENST00000398422.2 ENST00000537131.1 ENST00000538797.1 |
CCT7
|
chaperonin containing TCP1, subunit 7 (eta) |
| chr9_+_74920335 | 0.02 |
ENST00000451596.2
ENST00000436054.1 |
RP11-63P12.6
|
RP11-63P12.6 |
| chr2_+_73461364 | 0.02 |
ENST00000540468.1
ENST00000539919.1 ENST00000258091.5 |
CCT7
|
chaperonin containing TCP1, subunit 7 (eta) |
| chr3_+_49058444 | 0.02 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr9_+_74920408 | 0.02 |
ENST00000451152.1
|
RP11-63P12.6
|
RP11-63P12.6 |
| chr12_-_110937351 | 0.02 |
ENST00000552130.2
|
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr4_-_120225600 | 0.02 |
ENST00000399075.4
|
C4orf3
|
chromosome 4 open reading frame 3 |
| chr3_+_49057876 | 0.02 |
ENST00000326912.4
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr19_+_36157715 | 0.02 |
ENST00000379013.2
ENST00000222275.2 |
UPK1A
|
uroplakin 1A |
| chr3_-_10362725 | 0.01 |
ENST00000397109.3
ENST00000428626.1 ENST00000445064.1 ENST00000431352.1 ENST00000397117.1 ENST00000337354.4 ENST00000383801.2 ENST00000432213.1 ENST00000350697.3 |
SEC13
|
SEC13 homolog (S. cerevisiae) |
| chr15_-_60771280 | 0.01 |
ENST00000560072.1
ENST00000560406.1 ENST00000560520.1 ENST00000261520.4 ENST00000439632.1 |
NARG2
|
NMDA receptor regulated 2 |
| chr12_+_7941989 | 0.01 |
ENST00000229307.4
|
NANOG
|
Nanog homeobox |
| chr1_+_35734562 | 0.00 |
ENST00000314607.6
ENST00000373297.2 |
ZMYM4
|
zinc finger, MYM-type 4 |
| chr18_+_5748793 | 0.00 |
ENST00000566533.1
ENST00000562452.2 |
RP11-945C19.1
|
RP11-945C19.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DMBX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.0 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.1 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.0 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.1 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |