Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
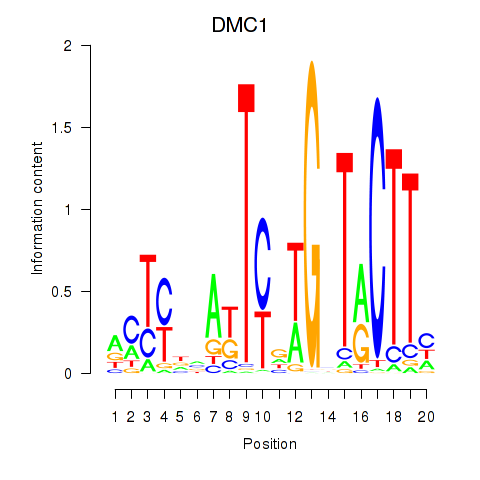
Results for DMC1
Z-value: 0.34
Transcription factors associated with DMC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DMC1
|
ENSG00000100206.5 | DNA meiotic recombinase 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DMC1 | hg19_v2_chr22_-_38966172_38966291 | 0.14 | 5.0e-01 | Click! |
Activity profile of DMC1 motif
Sorted Z-values of DMC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_76944621 | 2.03 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr17_+_77019030 | 0.61 |
ENST00000580454.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr17_+_77018896 | 0.60 |
ENST00000578229.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr7_+_142919130 | 0.59 |
ENST00000408947.3
|
TAS2R40
|
taste receptor, type 2, member 40 |
| chr22_+_21133469 | 0.44 |
ENST00000406799.1
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chrX_-_63450480 | 0.39 |
ENST00000362002.2
|
ASB12
|
ankyrin repeat and SOCS box containing 12 |
| chr17_+_31318886 | 0.35 |
ENST00000269053.3
ENST00000394638.1 |
SPACA3
|
sperm acrosome associated 3 |
| chr3_-_108672742 | 0.34 |
ENST00000261047.3
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr3_-_108672609 | 0.33 |
ENST00000393963.3
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr14_+_21525981 | 0.31 |
ENST00000308227.2
|
RNASE8
|
ribonuclease, RNase A family, 8 |
| chr2_-_190044480 | 0.31 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr3_+_101546827 | 0.25 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr5_-_36301984 | 0.24 |
ENST00000502994.1
ENST00000515759.1 ENST00000296604.3 |
RANBP3L
|
RAN binding protein 3-like |
| chr2_+_233497931 | 0.24 |
ENST00000264059.3
|
EFHD1
|
EF-hand domain family, member D1 |
| chr10_-_74856608 | 0.23 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chr11_-_85780086 | 0.22 |
ENST00000532317.1
ENST00000528256.1 ENST00000526033.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr11_-_85779971 | 0.21 |
ENST00000393346.3
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr6_+_26383318 | 0.20 |
ENST00000469230.1
ENST00000490025.1 ENST00000356709.4 ENST00000352867.2 ENST00000493275.1 ENST00000472507.1 ENST00000482536.1 ENST00000432533.2 ENST00000482842.1 |
BTN2A2
|
butyrophilin, subfamily 2, member A2 |
| chr21_-_46348694 | 0.19 |
ENST00000355153.4
ENST00000397850.2 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr6_-_62996066 | 0.19 |
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr2_+_187454749 | 0.19 |
ENST00000261023.3
ENST00000374907.3 |
ITGAV
|
integrin, alpha V |
| chr6_+_26383404 | 0.17 |
ENST00000416795.2
ENST00000494184.1 |
BTN2A2
|
butyrophilin, subfamily 2, member A2 |
| chr3_+_148457585 | 0.17 |
ENST00000402260.1
|
AGTR1
|
angiotensin II receptor, type 1 |
| chr18_-_5396271 | 0.17 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr21_-_46348626 | 0.17 |
ENST00000517563.1
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr15_-_42448788 | 0.16 |
ENST00000382396.4
ENST00000397272.3 |
PLA2G4F
|
phospholipase A2, group IVF |
| chr10_-_62332357 | 0.16 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr5_+_94727048 | 0.15 |
ENST00000283357.5
|
FAM81B
|
family with sequence similarity 81, member B |
| chr1_-_9714644 | 0.15 |
ENST00000377320.3
|
C1orf200
|
chromosome 1 open reading frame 200 |
| chr1_+_18958008 | 0.14 |
ENST00000420770.2
ENST00000400661.3 |
PAX7
|
paired box 7 |
| chr10_+_69865866 | 0.14 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr10_-_104597286 | 0.14 |
ENST00000369887.3
|
CYP17A1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr17_+_74075263 | 0.13 |
ENST00000334586.5
ENST00000392503.2 |
ZACN
|
zinc activated ligand-gated ion channel |
| chr7_-_22539771 | 0.12 |
ENST00000406890.2
ENST00000424363.1 |
STEAP1B
|
STEAP family member 1B |
| chr18_+_56892724 | 0.12 |
ENST00000456142.3
ENST00000530323.1 |
GRP
|
gastrin-releasing peptide |
| chr2_-_75426826 | 0.12 |
ENST00000305249.5
|
TACR1
|
tachykinin receptor 1 |
| chr11_+_71238313 | 0.11 |
ENST00000398536.4
|
KRTAP5-7
|
keratin associated protein 5-7 |
| chr6_-_31670723 | 0.11 |
ENST00000440843.2
ENST00000375842.4 |
ABHD16A
|
abhydrolase domain containing 16A |
| chr2_-_85641162 | 0.10 |
ENST00000447219.2
ENST00000409670.1 ENST00000409724.1 |
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr19_+_11466062 | 0.09 |
ENST00000251473.5
ENST00000591329.1 ENST00000586380.1 |
DKFZP761J1410
|
Lipid phosphate phosphatase-related protein type 2 |
| chr7_+_150758304 | 0.09 |
ENST00000482950.1
ENST00000463414.1 ENST00000310317.5 |
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr11_-_73720276 | 0.08 |
ENST00000348534.4
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr7_+_6793740 | 0.07 |
ENST00000403107.1
ENST00000404077.1 ENST00000435395.1 ENST00000418406.1 |
RSPH10B2
|
radial spoke head 10 homolog B2 (Chlamydomonas) |
| chr14_-_47351391 | 0.07 |
ENST00000399222.3
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr7_-_150884332 | 0.06 |
ENST00000275838.1
ENST00000422024.1 ENST00000434669.1 |
ASB10
|
ankyrin repeat and SOCS box containing 10 |
| chr7_-_6010263 | 0.06 |
ENST00000455618.2
ENST00000405415.1 ENST00000404406.1 ENST00000542644.1 |
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr6_+_116782527 | 0.06 |
ENST00000368606.3
ENST00000368605.1 |
FAM26F
|
family with sequence similarity 26, member F |
| chr6_+_46661575 | 0.06 |
ENST00000450697.1
|
TDRD6
|
tudor domain containing 6 |
| chr8_+_145438870 | 0.06 |
ENST00000527931.1
|
FAM203B
|
family with sequence similarity 203, member B |
| chr16_+_2255710 | 0.05 |
ENST00000397124.1
ENST00000565250.1 |
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr2_+_219247021 | 0.05 |
ENST00000539932.1
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr6_+_88054530 | 0.05 |
ENST00000388923.4
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chr10_+_11047259 | 0.05 |
ENST00000379261.4
ENST00000416382.2 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr17_-_60142609 | 0.05 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chr2_-_18741882 | 0.04 |
ENST00000381249.3
|
RDH14
|
retinol dehydrogenase 14 (all-trans/9-cis/11-cis) |
| chr17_+_38333263 | 0.04 |
ENST00000456989.2
ENST00000543876.1 ENST00000544503.1 ENST00000264644.6 ENST00000538884.1 |
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr19_-_42947121 | 0.04 |
ENST00000601181.1
|
CXCL17
|
chemokine (C-X-C motif) ligand 17 |
| chr2_-_163008903 | 0.04 |
ENST00000418842.2
ENST00000375497.3 |
GCG
|
glucagon |
| chr12_-_109025849 | 0.04 |
ENST00000228463.6
|
SELPLG
|
selectin P ligand |
| chr11_-_73720122 | 0.04 |
ENST00000426995.2
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chrX_-_134305322 | 0.04 |
ENST00000276241.6
ENST00000344129.2 |
CXorf48
|
cancer/testis antigen 55 |
| chr11_+_71846764 | 0.03 |
ENST00000456237.1
ENST00000442948.2 ENST00000546166.1 |
FOLR3
|
folate receptor 3 (gamma) |
| chr1_+_166958497 | 0.03 |
ENST00000367870.2
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr1_+_166958346 | 0.03 |
ENST00000367872.4
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr1_-_182642017 | 0.02 |
ENST00000367557.4
ENST00000258302.4 |
RGS8
|
regulator of G-protein signaling 8 |
| chr15_-_49103235 | 0.02 |
ENST00000380950.2
|
CEP152
|
centrosomal protein 152kDa |
| chr10_-_98031265 | 0.02 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chr14_-_21492113 | 0.02 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr7_-_150884873 | 0.02 |
ENST00000377867.3
|
ASB10
|
ankyrin repeat and SOCS box containing 10 |
| chr11_+_71846748 | 0.02 |
ENST00000445078.2
|
FOLR3
|
folate receptor 3 (gamma) |
| chr10_-_98031310 | 0.01 |
ENST00000427367.2
ENST00000413476.2 |
BLNK
|
B-cell linker |
| chr20_+_11008408 | 0.01 |
ENST00000378252.1
|
C20orf187
|
chromosome 20 open reading frame 187 |
| chr14_-_21492251 | 0.01 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chr10_-_135187193 | 0.00 |
ENST00000368547.3
|
ECHS1
|
enoyl CoA hydratase, short chain, 1, mitochondrial |
| chr1_-_190446759 | 0.00 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr20_+_123010 | 0.00 |
ENST00000382398.3
|
DEFB126
|
defensin, beta 126 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DMC1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.2 | 1.2 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.4 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.3 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.4 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.7 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.4 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.2 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.1 | GO:0035106 | angiotensin-mediated drinking behavior(GO:0003051) operant conditioning(GO:0035106) |
| 0.0 | 0.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.0 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.4 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.0 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.4 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.7 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.7 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.4 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.2 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.6 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 1.2 | GO:0005518 | collagen binding(GO:0005518) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |