Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
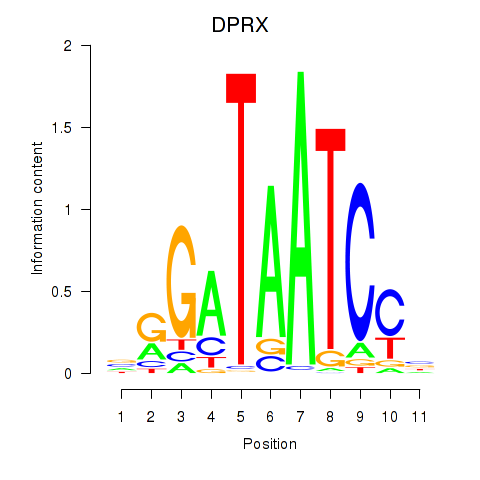
Results for DPRX
Z-value: 0.62
Transcription factors associated with DPRX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DPRX
|
ENSG00000204595.1 | divergent-paired related homeobox |
Activity profile of DPRX motif
Sorted Z-values of DPRX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_169079823 | 3.18 |
ENST00000367813.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr12_+_113344755 | 2.97 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr1_-_184943610 | 2.35 |
ENST00000367511.3
|
FAM129A
|
family with sequence similarity 129, member A |
| chr8_+_54764346 | 2.04 |
ENST00000297313.3
ENST00000344277.6 |
RGS20
|
regulator of G-protein signaling 20 |
| chr12_+_113344582 | 1.94 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr1_-_153521597 | 1.77 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr1_-_150208320 | 1.64 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr1_-_153521714 | 1.57 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr17_+_77021702 | 1.40 |
ENST00000392445.2
ENST00000354124.3 |
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr12_+_113344811 | 1.16 |
ENST00000551241.1
ENST00000553185.1 ENST00000550689.1 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr5_+_35856951 | 1.14 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr6_-_35480640 | 1.07 |
ENST00000428978.1
ENST00000322263.4 |
TULP1
|
tubby like protein 1 |
| chr4_+_89299885 | 1.07 |
ENST00000380265.5
ENST00000273960.3 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr1_+_15986364 | 0.97 |
ENST00000345034.1
|
RSC1A1
|
regulatory solute carrier protein, family 1, member 1 |
| chr1_-_173174681 | 0.95 |
ENST00000367718.1
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr7_+_28452130 | 0.89 |
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr6_-_37225391 | 0.88 |
ENST00000356757.2
|
TMEM217
|
transmembrane protein 217 |
| chr6_-_37225367 | 0.87 |
ENST00000336655.2
|
TMEM217
|
transmembrane protein 217 |
| chr8_+_104384616 | 0.85 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr5_-_150460914 | 0.77 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr11_-_104905840 | 0.72 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr2_+_232063436 | 0.65 |
ENST00000440107.1
|
ARMC9
|
armadillo repeat containing 9 |
| chr6_+_31554826 | 0.63 |
ENST00000376089.2
ENST00000396112.2 |
LST1
|
leukocyte specific transcript 1 |
| chr6_+_31555045 | 0.59 |
ENST00000396101.3
ENST00000490742.1 |
LST1
|
leukocyte specific transcript 1 |
| chr11_+_61957687 | 0.57 |
ENST00000306238.3
|
SCGB1D1
|
secretoglobin, family 1D, member 1 |
| chr6_+_26383404 | 0.57 |
ENST00000416795.2
ENST00000494184.1 |
BTN2A2
|
butyrophilin, subfamily 2, member A2 |
| chr9_+_109625378 | 0.57 |
ENST00000277225.5
ENST00000457913.1 ENST00000472574.1 |
ZNF462
|
zinc finger protein 462 |
| chr11_+_61248583 | 0.56 |
ENST00000432063.2
ENST00000338608.2 |
PPP1R32
|
protein phosphatase 1, regulatory subunit 32 |
| chr16_-_74734742 | 0.54 |
ENST00000308807.7
ENST00000573267.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr6_+_4706368 | 0.54 |
ENST00000328908.5
|
CDYL
|
chromodomain protein, Y-like |
| chr5_-_124081008 | 0.53 |
ENST00000306315.5
|
ZNF608
|
zinc finger protein 608 |
| chr6_-_35480705 | 0.53 |
ENST00000229771.6
|
TULP1
|
tubby like protein 1 |
| chr6_-_33290580 | 0.49 |
ENST00000446511.1
ENST00000446403.1 ENST00000414083.2 ENST00000266000.6 ENST00000374542.5 |
DAXX
|
death-domain associated protein |
| chr9_-_22009297 | 0.49 |
ENST00000276925.6
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| chr10_+_74451883 | 0.48 |
ENST00000373053.3
ENST00000357157.6 |
MCU
|
mitochondrial calcium uniporter |
| chr4_-_170924888 | 0.48 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr1_-_150780757 | 0.47 |
ENST00000271651.3
|
CTSK
|
cathepsin K |
| chr1_+_50569575 | 0.46 |
ENST00000371827.1
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr2_-_49381646 | 0.45 |
ENST00000346173.3
ENST00000406846.2 |
FSHR
|
follicle stimulating hormone receptor |
| chr2_-_49381572 | 0.45 |
ENST00000454032.1
ENST00000304421.4 |
FSHR
|
follicle stimulating hormone receptor |
| chr6_+_26383318 | 0.44 |
ENST00000469230.1
ENST00000490025.1 ENST00000356709.4 ENST00000352867.2 ENST00000493275.1 ENST00000472507.1 ENST00000482536.1 ENST00000432533.2 ENST00000482842.1 |
BTN2A2
|
butyrophilin, subfamily 2, member A2 |
| chr1_+_179561011 | 0.43 |
ENST00000294848.8
ENST00000444136.1 |
TDRD5
|
tudor domain containing 5 |
| chr5_-_59064458 | 0.42 |
ENST00000502575.1
ENST00000507116.1 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr6_+_7108210 | 0.42 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr20_+_361890 | 0.39 |
ENST00000449710.1
ENST00000422053.2 |
TRIB3
|
tribbles pseudokinase 3 |
| chr1_+_20396649 | 0.38 |
ENST00000375108.3
|
PLA2G5
|
phospholipase A2, group V |
| chr16_-_30022735 | 0.38 |
ENST00000564944.1
|
DOC2A
|
double C2-like domains, alpha |
| chr6_+_37225540 | 0.38 |
ENST00000373491.3
|
TBC1D22B
|
TBC1 domain family, member 22B |
| chrX_+_153448107 | 0.37 |
ENST00000369935.5
|
OPN1MW
|
opsin 1 (cone pigments), medium-wave-sensitive |
| chr12_+_113416265 | 0.36 |
ENST00000449768.2
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr1_-_170043709 | 0.36 |
ENST00000367767.1
ENST00000361580.2 ENST00000538366.1 |
KIFAP3
|
kinesin-associated protein 3 |
| chr4_-_103997862 | 0.35 |
ENST00000394785.3
|
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr12_-_71182695 | 0.35 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr6_-_132967142 | 0.34 |
ENST00000275216.1
|
TAAR1
|
trace amine associated receptor 1 |
| chr19_+_36132631 | 0.33 |
ENST00000379026.2
ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2
|
ets variant 2 |
| chr3_+_111393659 | 0.33 |
ENST00000477665.1
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr11_-_104972158 | 0.33 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr18_-_64271363 | 0.33 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr3_-_112564797 | 0.33 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr7_+_134464376 | 0.33 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr16_-_74734672 | 0.33 |
ENST00000306247.7
ENST00000575686.1 |
MLKL
|
mixed lineage kinase domain-like |
| chrX_+_15767971 | 0.31 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr15_+_63481668 | 0.31 |
ENST00000321437.4
ENST00000559006.1 ENST00000448330.2 |
RAB8B
|
RAB8B, member RAS oncogene family |
| chr3_+_130650738 | 0.31 |
ENST00000504612.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr7_+_134576317 | 0.31 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr20_+_8112824 | 0.31 |
ENST00000378641.3
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific) |
| chr9_+_37667978 | 0.31 |
ENST00000539465.1
|
FRMPD1
|
FERM and PDZ domain containing 1 |
| chr5_+_140345820 | 0.31 |
ENST00000289269.5
|
PCDHAC2
|
protocadherin alpha subfamily C, 2 |
| chr3_+_63953415 | 0.30 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr19_+_36157715 | 0.30 |
ENST00000379013.2
ENST00000222275.2 |
UPK1A
|
uroplakin 1A |
| chr4_-_103998060 | 0.30 |
ENST00000339611.4
|
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr2_+_234216454 | 0.30 |
ENST00000447536.1
ENST00000409110.1 |
SAG
|
S-antigen; retina and pineal gland (arrestin) |
| chr4_-_103998439 | 0.29 |
ENST00000503230.1
ENST00000503818.1 |
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr11_-_85430204 | 0.29 |
ENST00000389958.3
ENST00000527794.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr11_-_85430163 | 0.28 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr3_+_111393501 | 0.28 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr11_-_85430088 | 0.28 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr5_+_63461642 | 0.28 |
ENST00000296615.6
ENST00000381081.2 ENST00000389100.4 |
RNF180
|
ring finger protein 180 |
| chr11_+_120973375 | 0.28 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr2_-_235405168 | 0.28 |
ENST00000339728.3
|
ARL4C
|
ADP-ribosylation factor-like 4C |
| chr1_+_12916941 | 0.28 |
ENST00000240189.2
|
PRAMEF2
|
PRAME family member 2 |
| chr12_-_113574028 | 0.26 |
ENST00000546530.1
ENST00000261729.5 |
RASAL1
|
RAS protein activator like 1 (GAP1 like) |
| chr7_+_134576151 | 0.26 |
ENST00000393118.2
|
CALD1
|
caldesmon 1 |
| chr18_-_64271316 | 0.26 |
ENST00000540086.1
ENST00000580157.1 |
CDH19
|
cadherin 19, type 2 |
| chr7_+_134464414 | 0.26 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr1_-_13452656 | 0.26 |
ENST00000376132.3
|
PRAMEF13
|
PRAME family member 13 |
| chr2_+_69201705 | 0.25 |
ENST00000377938.2
|
GKN1
|
gastrokine 1 |
| chr2_+_27799389 | 0.25 |
ENST00000408964.2
|
C2orf16
|
chromosome 2 open reading frame 16 |
| chr1_+_183441618 | 0.25 |
ENST00000507691.2
ENST00000508461.1 ENST00000419169.1 ENST00000347615.2 ENST00000507469.1 ENST00000515829.2 |
SMG7
|
SMG7 nonsense mediated mRNA decay factor |
| chr1_+_16767167 | 0.25 |
ENST00000337132.5
|
NECAP2
|
NECAP endocytosis associated 2 |
| chr2_-_30144432 | 0.25 |
ENST00000389048.3
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase |
| chr1_+_183441500 | 0.25 |
ENST00000456731.2
|
SMG7
|
SMG7 nonsense mediated mRNA decay factor |
| chr3_+_148447887 | 0.25 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr17_-_42019836 | 0.25 |
ENST00000225992.3
|
PPY
|
pancreatic polypeptide |
| chr17_-_15244894 | 0.24 |
ENST00000338696.2
ENST00000543896.1 ENST00000539245.1 ENST00000539316.1 ENST00000395930.1 |
TEKT3
|
tektin 3 |
| chr19_-_59070239 | 0.24 |
ENST00000595957.1
ENST00000253023.3 |
UBE2M
|
ubiquitin-conjugating enzyme E2M |
| chr15_+_34261089 | 0.24 |
ENST00000383263.5
|
CHRM5
|
cholinergic receptor, muscarinic 5 |
| chr11_-_62752162 | 0.23 |
ENST00000458333.2
ENST00000421062.2 |
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr1_+_183441600 | 0.23 |
ENST00000367537.3
|
SMG7
|
SMG7 nonsense mediated mRNA decay factor |
| chr14_-_101034407 | 0.23 |
ENST00000443071.2
ENST00000557378.1 |
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr10_-_72362515 | 0.23 |
ENST00000373209.2
ENST00000441259.1 |
PRF1
|
perforin 1 (pore forming protein) |
| chr8_-_71520513 | 0.23 |
ENST00000262213.2
ENST00000536748.1 ENST00000518678.1 |
TRAM1
|
translocation associated membrane protein 1 |
| chr1_-_27481401 | 0.22 |
ENST00000263980.3
|
SLC9A1
|
solute carrier family 9, subfamily A (NHE1, cation proton antiporter 1), member 1 |
| chr12_+_124069070 | 0.22 |
ENST00000262225.3
ENST00000438031.2 |
TMED2
|
transmembrane emp24 domain trafficking protein 2 |
| chr9_-_132404374 | 0.21 |
ENST00000277459.4
ENST00000450050.2 ENST00000277458.4 |
ASB6
|
ankyrin repeat and SOCS box containing 6 |
| chr5_+_169010638 | 0.21 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr19_+_8455077 | 0.21 |
ENST00000328024.6
|
RAB11B
|
RAB11B, member RAS oncogene family |
| chr6_-_32908765 | 0.21 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr6_-_26108355 | 0.21 |
ENST00000338379.4
|
HIST1H1T
|
histone cluster 1, H1t |
| chr6_+_36164487 | 0.20 |
ENST00000357641.6
|
BRPF3
|
bromodomain and PHD finger containing, 3 |
| chr4_-_122791583 | 0.20 |
ENST00000506636.1
ENST00000264499.4 |
BBS7
|
Bardet-Biedl syndrome 7 |
| chr12_-_110937351 | 0.20 |
ENST00000552130.2
|
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr22_-_43355858 | 0.20 |
ENST00000402229.1
ENST00000407585.1 ENST00000453079.1 |
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr16_+_21244986 | 0.20 |
ENST00000311620.5
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr17_+_3118915 | 0.20 |
ENST00000304094.1
|
OR1A1
|
olfactory receptor, family 1, subfamily A, member 1 |
| chr10_-_89577910 | 0.20 |
ENST00000308448.7
ENST00000541004.1 |
ATAD1
|
ATPase family, AAA domain containing 1 |
| chr9_-_123239632 | 0.20 |
ENST00000416449.1
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr9_+_4839762 | 0.20 |
ENST00000448872.2
ENST00000441844.1 |
RCL1
|
RNA terminal phosphate cyclase-like 1 |
| chr11_-_104817919 | 0.19 |
ENST00000533252.1
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr19_-_12267524 | 0.19 |
ENST00000455799.1
ENST00000355738.1 ENST00000439556.2 ENST00000542938.1 |
ZNF625
|
zinc finger protein 625 |
| chr18_+_32290218 | 0.19 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
| chr1_+_111415757 | 0.19 |
ENST00000429072.2
ENST00000271324.5 |
CD53
|
CD53 molecule |
| chr16_+_81272287 | 0.19 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr11_-_30608413 | 0.19 |
ENST00000528686.1
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chr12_+_54891495 | 0.18 |
ENST00000293373.6
|
NCKAP1L
|
NCK-associated protein 1-like |
| chr2_-_190044480 | 0.18 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr6_+_7107999 | 0.18 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr1_+_16767195 | 0.18 |
ENST00000504551.2
ENST00000457722.2 ENST00000406746.1 ENST00000443980.2 |
NECAP2
|
NECAP endocytosis associated 2 |
| chr6_-_22297730 | 0.18 |
ENST00000306482.1
|
PRL
|
prolactin |
| chr6_-_32908792 | 0.18 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr1_+_182808474 | 0.18 |
ENST00000367549.3
|
DHX9
|
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr5_+_180336564 | 0.18 |
ENST00000505126.1
ENST00000533815.2 |
BTNL8
|
butyrophilin-like 8 |
| chr17_-_9694614 | 0.18 |
ENST00000330255.5
ENST00000571134.1 |
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C |
| chr19_-_50370799 | 0.18 |
ENST00000600910.1
ENST00000322344.3 ENST00000600573.1 |
PNKP
|
polynucleotide kinase 3'-phosphatase |
| chr17_+_55183261 | 0.17 |
ENST00000576295.1
|
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr17_+_7531281 | 0.17 |
ENST00000575729.1
ENST00000340624.5 |
SHBG
|
sex hormone-binding globulin |
| chr3_-_124653579 | 0.17 |
ENST00000478191.1
ENST00000311075.3 |
MUC13
|
mucin 13, cell surface associated |
| chr19_+_50528971 | 0.17 |
ENST00000598809.1
ENST00000595661.1 ENST00000391821.2 |
ZNF473
|
zinc finger protein 473 |
| chr3_+_133502877 | 0.17 |
ENST00000466490.2
|
SRPRB
|
signal recognition particle receptor, B subunit |
| chr1_-_236046872 | 0.16 |
ENST00000536965.1
|
LYST
|
lysosomal trafficking regulator |
| chr7_+_123241908 | 0.16 |
ENST00000434204.1
ENST00000437535.1 ENST00000451215.1 |
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chrX_+_69488174 | 0.16 |
ENST00000480877.2
ENST00000307959.8 |
ARR3
|
arrestin 3, retinal (X-arrestin) |
| chr18_-_3845293 | 0.15 |
ENST00000400145.2
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr21_-_28217721 | 0.15 |
ENST00000284984.3
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr2_-_180610767 | 0.15 |
ENST00000409343.1
|
ZNF385B
|
zinc finger protein 385B |
| chr12_+_104359641 | 0.15 |
ENST00000537100.1
|
TDG
|
thymine-DNA glycosylase |
| chr11_-_65629497 | 0.15 |
ENST00000532134.1
|
CFL1
|
cofilin 1 (non-muscle) |
| chrX_+_118425471 | 0.14 |
ENST00000428222.1
|
RP5-1139I1.1
|
RP5-1139I1.1 |
| chr16_+_30386098 | 0.14 |
ENST00000322861.7
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr8_-_145701718 | 0.14 |
ENST00000377317.4
|
FOXH1
|
forkhead box H1 |
| chr8_+_65285851 | 0.14 |
ENST00000520799.1
ENST00000521441.1 |
LINC00966
|
long intergenic non-protein coding RNA 966 |
| chr2_+_113342011 | 0.14 |
ENST00000324913.5
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr19_+_56165480 | 0.14 |
ENST00000450554.2
|
U2AF2
|
U2 small nuclear RNA auxiliary factor 2 |
| chr8_-_79717750 | 0.14 |
ENST00000263851.4
ENST00000379113.2 |
IL7
|
interleukin 7 |
| chr11_-_111944895 | 0.14 |
ENST00000431456.1
ENST00000280350.4 ENST00000530641.1 |
PIH1D2
|
PIH1 domain containing 2 |
| chr9_-_123812542 | 0.14 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr19_-_50529193 | 0.14 |
ENST00000596445.1
ENST00000599538.1 |
VRK3
|
vaccinia related kinase 3 |
| chr13_+_52436111 | 0.14 |
ENST00000242819.4
|
CCDC70
|
coiled-coil domain containing 70 |
| chr9_-_95244781 | 0.14 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr11_+_12399071 | 0.14 |
ENST00000539723.1
ENST00000550549.1 |
PARVA
|
parvin, alpha |
| chr2_-_26700900 | 0.14 |
ENST00000338581.6
ENST00000339598.3 ENST00000402415.3 |
OTOF
|
otoferlin |
| chr2_+_113342163 | 0.14 |
ENST00000409719.1
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr1_+_156117149 | 0.13 |
ENST00000435124.1
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr3_+_77088989 | 0.13 |
ENST00000461745.1
|
ROBO2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr14_-_21056121 | 0.13 |
ENST00000557105.1
ENST00000398008.2 ENST00000555841.1 ENST00000443456.2 ENST00000432835.2 ENST00000557503.1 ENST00000398009.2 ENST00000554842.1 |
RNASE11
|
ribonuclease, RNase A family, 11 (non-active) |
| chr16_-_46723066 | 0.13 |
ENST00000299138.7
|
VPS35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chrX_+_71996972 | 0.13 |
ENST00000334036.5
|
DMRTC1B
|
DMRT-like family C1B |
| chr7_-_135412925 | 0.13 |
ENST00000354042.4
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr15_+_75970672 | 0.13 |
ENST00000435356.1
|
AC105020.1
|
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chr18_-_28681950 | 0.13 |
ENST00000251081.6
|
DSC2
|
desmocollin 2 |
| chr12_+_104359614 | 0.12 |
ENST00000266775.9
ENST00000544861.1 |
TDG
|
thymine-DNA glycosylase |
| chr18_-_28682374 | 0.12 |
ENST00000280904.6
|
DSC2
|
desmocollin 2 |
| chr7_-_30066233 | 0.12 |
ENST00000222803.5
|
FKBP14
|
FK506 binding protein 14, 22 kDa |
| chr4_-_152149033 | 0.12 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr1_+_100435315 | 0.12 |
ENST00000370155.3
ENST00000465289.1 |
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr11_-_10715287 | 0.12 |
ENST00000423302.2
|
MRVI1
|
murine retrovirus integration site 1 homolog |
| chr22_-_19137796 | 0.12 |
ENST00000086933.2
|
GSC2
|
goosecoid homeobox 2 |
| chr14_-_21055874 | 0.12 |
ENST00000553849.1
|
RNASE11
|
ribonuclease, RNase A family, 11 (non-active) |
| chr4_-_153700864 | 0.12 |
ENST00000304337.2
|
TIGD4
|
tigger transposable element derived 4 |
| chr2_+_145425573 | 0.12 |
ENST00000600064.1
ENST00000597670.1 ENST00000414256.1 ENST00000599187.1 ENST00000451774.1 ENST00000599072.1 ENST00000596589.1 ENST00000597893.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chrX_-_53449593 | 0.12 |
ENST00000375340.6
ENST00000322213.4 |
SMC1A
|
structural maintenance of chromosomes 1A |
| chr4_-_47465666 | 0.11 |
ENST00000381571.4
|
COMMD8
|
COMM domain containing 8 |
| chr3_+_141105705 | 0.11 |
ENST00000513258.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr10_+_51572408 | 0.11 |
ENST00000374082.1
|
NCOA4
|
nuclear receptor coactivator 4 |
| chr17_-_48785216 | 0.11 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr6_-_30640811 | 0.11 |
ENST00000376442.3
|
DHX16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr12_+_6949964 | 0.11 |
ENST00000541978.1
ENST00000435982.2 |
GNB3
|
guanine nucleotide binding protein (G protein), beta polypeptide 3 |
| chr19_-_57967854 | 0.11 |
ENST00000321039.3
|
VN1R1
|
vomeronasal 1 receptor 1 |
| chr1_+_236557569 | 0.11 |
ENST00000334232.4
|
EDARADD
|
EDAR-associated death domain |
| chr3_-_54962100 | 0.11 |
ENST00000273286.5
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr7_-_75401513 | 0.11 |
ENST00000005180.4
|
CCL26
|
chemokine (C-C motif) ligand 26 |
| chr4_+_84377198 | 0.11 |
ENST00000507349.1
ENST00000509970.1 ENST00000505719.1 |
MRPS18C
|
mitochondrial ribosomal protein S18C |
| chr9_-_22009241 | 0.11 |
ENST00000380142.4
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| chr2_-_119605253 | 0.11 |
ENST00000295206.6
|
EN1
|
engrailed homeobox 1 |
| chr10_+_51572339 | 0.11 |
ENST00000344348.6
|
NCOA4
|
nuclear receptor coactivator 4 |
| chr5_-_150948414 | 0.11 |
ENST00000261800.5
|
FAT2
|
FAT atypical cadherin 2 |
| chr5_+_150040403 | 0.10 |
ENST00000517768.1
ENST00000297130.4 |
MYOZ3
|
myozenin 3 |
| chr3_+_136676851 | 0.10 |
ENST00000309741.5
|
IL20RB
|
interleukin 20 receptor beta |
| chr6_-_31080336 | 0.10 |
ENST00000259870.3
|
C6orf15
|
chromosome 6 open reading frame 15 |
| chr15_+_81475047 | 0.10 |
ENST00000559388.1
|
IL16
|
interleukin 16 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DPRX
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.4 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.3 | 0.9 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.3 | 1.0 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.2 | 0.9 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.2 | 0.8 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.2 | 0.6 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.2 | 1.4 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 1.6 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 0.6 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.4 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 0.5 | GO:1903935 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.1 | 0.8 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 0.4 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 | 0.9 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.3 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 0.1 | 0.4 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 1.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.3 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.1 | 0.2 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.1 | 0.8 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.5 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 0.3 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.3 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.9 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.3 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 0.2 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) negative regulation of hypersensitivity(GO:0002884) |
| 0.0 | 0.1 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.4 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.0 | 0.3 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:1903181 | positive regulation of Wnt protein secretion(GO:0061357) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.0 | 5.9 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.1 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.2 | GO:0035546 | interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 0.0 | 0.2 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.4 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 1.6 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.2 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.9 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:2000814 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.2 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) visceral serous pericardium development(GO:0061032) |
| 0.0 | 0.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.6 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.1 | GO:0072396 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.4 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.2 | GO:0002418 | immune response to tumor cell(GO:0002418) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0051105 | regulation of DNA ligation(GO:0051105) |
| 0.0 | 1.2 | GO:0050672 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 2.0 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 2.0 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.0 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.1 | 3.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.4 | GO:0016939 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.1 | 1.7 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 1.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.5 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 0.3 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.4 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 2.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.3 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.9 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.2 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 2.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.4 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.4 | 1.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.2 | 0.9 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.2 | 0.9 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.2 | 3.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.5 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.3 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.2 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 0.2 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) DNA/RNA helicase activity(GO:0033677) |
| 0.1 | 1.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.2 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.3 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.2 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.2 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 1.6 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.2 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.4 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 1.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.7 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.0 | GO:0008200 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.1 | GO:0016295 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.0 | 0.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 2.3 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 3.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 6.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 0.9 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.5 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 2.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |