Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
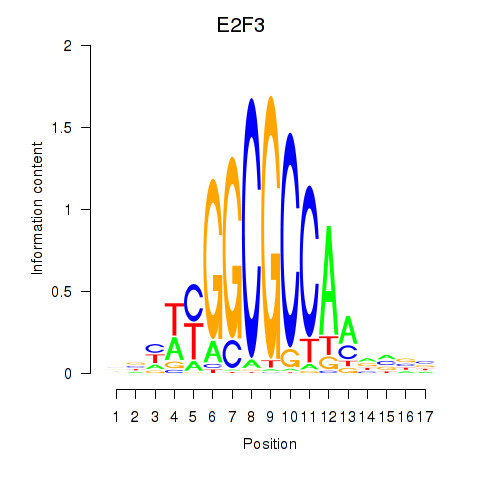
Results for E2F3
Z-value: 1.41
Transcription factors associated with E2F3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2F3
|
ENSG00000112242.10 | E2F transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2F3 | hg19_v2_chr6_+_20403997_20404034 | 0.04 | 8.6e-01 | Click! |
Activity profile of E2F3 motif
Sorted Z-values of E2F3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_89841024 | 4.26 |
ENST00000394626.1
|
STEAP2
|
STEAP family member 2, metalloreductase |
| chr9_-_39288092 | 4.15 |
ENST00000323947.7
ENST00000297668.6 ENST00000377656.2 ENST00000377659.1 |
CNTNAP3
|
contactin associated protein-like 3 |
| chr8_-_6836156 | 3.96 |
ENST00000382679.2
|
DEFA1
|
defensin, alpha 1 |
| chr7_+_89841000 | 3.79 |
ENST00000287908.3
|
STEAP2
|
STEAP family member 2, metalloreductase |
| chr7_+_89841144 | 3.73 |
ENST00000394622.2
ENST00000394632.1 ENST00000426158.1 ENST00000394621.2 ENST00000402625.2 |
STEAP2
|
STEAP family member 2, metalloreductase |
| chr9_+_79074068 | 3.49 |
ENST00000444201.2
ENST00000376730.4 |
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr16_-_88772761 | 3.48 |
ENST00000567844.1
ENST00000312838.4 |
RNF166
|
ring finger protein 166 |
| chr20_-_23030296 | 3.39 |
ENST00000377103.2
|
THBD
|
thrombomodulin |
| chr2_+_112656048 | 3.37 |
ENST00000295408.4
|
MERTK
|
c-mer proto-oncogene tyrosine kinase |
| chr8_-_81083341 | 3.31 |
ENST00000519303.2
|
TPD52
|
tumor protein D52 |
| chr14_-_91526462 | 3.29 |
ENST00000536315.2
|
RPS6KA5
|
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
| chr16_-_88772670 | 3.07 |
ENST00000562544.1
|
RNF166
|
ring finger protein 166 |
| chr12_-_42877726 | 2.92 |
ENST00000548696.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr1_+_77997785 | 2.88 |
ENST00000478255.1
|
AK5
|
adenylate kinase 5 |
| chr1_+_61547405 | 2.82 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr2_+_112656176 | 2.65 |
ENST00000421804.2
ENST00000409780.1 |
MERTK
|
c-mer proto-oncogene tyrosine kinase |
| chr5_-_100238956 | 2.63 |
ENST00000231461.5
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr4_-_18023350 | 2.54 |
ENST00000539056.1
ENST00000382226.5 ENST00000326877.4 |
LCORL
|
ligand dependent nuclear receptor corepressor-like |
| chr12_-_42877408 | 2.52 |
ENST00000552240.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr15_-_57025759 | 2.50 |
ENST00000267807.7
|
ZNF280D
|
zinc finger protein 280D |
| chr3_+_39851094 | 2.43 |
ENST00000302541.6
|
MYRIP
|
myosin VIIA and Rab interacting protein |
| chr5_-_111093406 | 2.42 |
ENST00000379671.3
|
NREP
|
neuronal regeneration related protein |
| chr12_-_42877764 | 2.39 |
ENST00000455697.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr12_-_42983478 | 2.35 |
ENST00000345127.3
ENST00000547113.1 |
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr1_+_25944341 | 2.35 |
ENST00000263979.3
|
MAN1C1
|
mannosidase, alpha, class 1C, member 1 |
| chr18_+_11981427 | 2.34 |
ENST00000269159.3
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr9_-_77703115 | 2.25 |
ENST00000361092.4
ENST00000376808.4 |
NMRK1
|
nicotinamide riboside kinase 1 |
| chr1_+_77747656 | 2.23 |
ENST00000354567.2
|
AK5
|
adenylate kinase 5 |
| chr9_-_111775772 | 2.21 |
ENST00000325580.6
ENST00000374593.4 ENST00000374595.4 ENST00000325551.4 |
CTNNAL1
|
catenin (cadherin-associated protein), alpha-like 1 |
| chr6_+_142623063 | 2.21 |
ENST00000296932.8
ENST00000367609.3 |
GPR126
|
G protein-coupled receptor 126 |
| chr1_-_211752073 | 2.18 |
ENST00000367001.4
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1 |
| chr13_-_110438914 | 2.00 |
ENST00000375856.3
|
IRS2
|
insulin receptor substrate 2 |
| chr4_-_123844084 | 2.00 |
ENST00000339154.2
|
NUDT6
|
nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
| chr7_+_12727250 | 1.98 |
ENST00000404894.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr16_+_14165160 | 1.96 |
ENST00000574998.1
ENST00000571589.1 ENST00000574045.1 |
MKL2
|
MKL/myocardin-like 2 |
| chr18_+_77623668 | 1.96 |
ENST00000316249.3
|
KCNG2
|
potassium voltage-gated channel, subfamily G, member 2 |
| chr1_+_61548225 | 1.95 |
ENST00000371187.3
|
NFIA
|
nuclear factor I/A |
| chrX_-_125686784 | 1.91 |
ENST00000371126.1
|
DCAF12L1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chrX_+_53123314 | 1.87 |
ENST00000605526.1
ENST00000604062.1 ENST00000604369.1 ENST00000366185.2 ENST00000604849.1 |
RP11-258C19.5
|
long intergenic non-protein coding RNA 1155 |
| chrX_+_46433193 | 1.87 |
ENST00000276055.3
|
CHST7
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
| chr12_-_95044309 | 1.86 |
ENST00000261226.4
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr19_-_35454953 | 1.84 |
ENST00000404801.1
|
ZNF792
|
zinc finger protein 792 |
| chr7_-_21985489 | 1.84 |
ENST00000356195.5
ENST00000447180.1 ENST00000373934.4 ENST00000457951.1 |
CDCA7L
|
cell division cycle associated 7-like |
| chr2_-_39664405 | 1.84 |
ENST00000341681.5
ENST00000263881.3 |
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr8_+_96037255 | 1.77 |
ENST00000286687.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr2_-_75788038 | 1.77 |
ENST00000393913.3
ENST00000410113.1 |
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr9_-_77703056 | 1.73 |
ENST00000376811.1
|
NMRK1
|
nicotinamide riboside kinase 1 |
| chr7_-_86974767 | 1.72 |
ENST00000610086.1
|
TP53TG1
|
TP53 target 1 (non-protein coding) |
| chr8_+_56792377 | 1.71 |
ENST00000520220.2
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
| chr3_+_110790590 | 1.70 |
ENST00000485303.1
|
PVRL3
|
poliovirus receptor-related 3 |
| chr7_-_86974785 | 1.70 |
ENST00000432193.1
ENST00000421293.1 ENST00000542586.1 ENST00000359941.5 ENST00000416560.1 |
TP53TG1
|
TP53 target 1 (non-protein coding) |
| chr7_+_12726474 | 1.69 |
ENST00000396662.1
ENST00000356797.3 ENST00000396664.2 |
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr18_+_59992527 | 1.67 |
ENST00000586569.1
|
TNFRSF11A
|
tumor necrosis factor receptor superfamily, member 11a, NFKB activator |
| chr6_-_27440837 | 1.66 |
ENST00000211936.6
|
ZNF184
|
zinc finger protein 184 |
| chr6_-_84419101 | 1.63 |
ENST00000520302.1
ENST00000520213.1 ENST00000439399.2 ENST00000428679.2 ENST00000437520.1 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr4_+_87928413 | 1.63 |
ENST00000544085.1
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr5_-_111093759 | 1.61 |
ENST00000509979.1
ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP
|
neuronal regeneration related protein |
| chr15_+_83776324 | 1.61 |
ENST00000379390.6
ENST00000379386.4 ENST00000565774.1 ENST00000565982.1 |
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr1_+_2160134 | 1.60 |
ENST00000378536.4
|
SKI
|
v-ski avian sarcoma viral oncogene homolog |
| chr3_+_32147997 | 1.60 |
ENST00000282541.5
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chrX_+_13671225 | 1.60 |
ENST00000545566.1
ENST00000544987.1 ENST00000314720.4 |
TCEANC
|
transcription elongation factor A (SII) N-terminal and central domain containing |
| chr10_-_131762105 | 1.57 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr8_+_96037205 | 1.56 |
ENST00000396124.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr1_+_53068020 | 1.55 |
ENST00000361314.4
|
GPX7
|
glutathione peroxidase 7 |
| chr5_-_111092930 | 1.55 |
ENST00000257435.7
|
NREP
|
neuronal regeneration related protein |
| chr1_+_63249796 | 1.52 |
ENST00000443289.1
ENST00000317868.4 ENST00000371120.3 |
ATG4C
|
autophagy related 4C, cysteine peptidase |
| chr8_-_52811714 | 1.51 |
ENST00000544451.1
|
PCMTD1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr14_-_30396948 | 1.50 |
ENST00000331968.5
|
PRKD1
|
protein kinase D1 |
| chr5_-_79551838 | 1.50 |
ENST00000509193.1
ENST00000512972.2 |
SERINC5
|
serine incorporator 5 |
| chr6_+_34857019 | 1.50 |
ENST00000360359.3
ENST00000535627.1 |
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A |
| chr6_-_144385698 | 1.50 |
ENST00000444202.1
ENST00000437412.1 |
PLAGL1
|
pleiomorphic adenoma gene-like 1 |
| chr16_-_56553882 | 1.48 |
ENST00000568104.1
|
BBS2
|
Bardet-Biedl syndrome 2 |
| chr2_+_8822113 | 1.48 |
ENST00000396290.1
ENST00000331129.3 |
ID2
|
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein |
| chr14_+_77228532 | 1.45 |
ENST00000167106.4
ENST00000554237.1 |
VASH1
|
vasohibin 1 |
| chr10_+_35415719 | 1.45 |
ENST00000474362.1
ENST00000374721.3 |
CREM
|
cAMP responsive element modulator |
| chr12_-_15942503 | 1.44 |
ENST00000281172.5
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr19_-_14628645 | 1.44 |
ENST00000598235.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr14_-_75536182 | 1.42 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr15_+_83776137 | 1.42 |
ENST00000322019.9
|
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr1_-_59043166 | 1.41 |
ENST00000371225.2
|
TACSTD2
|
tumor-associated calcium signal transducer 2 |
| chr14_-_30396802 | 1.41 |
ENST00000415220.2
|
PRKD1
|
protein kinase D1 |
| chr2_-_70780572 | 1.39 |
ENST00000450929.1
|
TGFA
|
transforming growth factor, alpha |
| chr4_+_87928140 | 1.39 |
ENST00000307808.6
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr11_-_105948129 | 1.38 |
ENST00000526793.1
|
KBTBD3
|
kelch repeat and BTB (POZ) domain containing 3 |
| chr15_+_66585555 | 1.38 |
ENST00000319194.5
ENST00000525134.2 ENST00000441424.2 |
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr5_-_111092873 | 1.38 |
ENST00000509025.1
ENST00000515855.1 |
NREP
|
neuronal regeneration related protein |
| chr12_-_22697343 | 1.36 |
ENST00000446597.1
ENST00000536386.1 ENST00000396028.2 ENST00000545552.1 ENST00000544930.1 ENST00000333957.4 |
C2CD5
|
C2 calcium-dependent domain containing 5 |
| chr2_+_181845298 | 1.34 |
ENST00000410062.4
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr2_-_85829780 | 1.34 |
ENST00000334462.5
|
TMEM150A
|
transmembrane protein 150A |
| chr19_-_49552006 | 1.34 |
ENST00000391869.3
|
CGB1
|
chorionic gonadotropin, beta polypeptide 1 |
| chr16_+_66586461 | 1.33 |
ENST00000264001.4
ENST00000351137.4 ENST00000345436.4 ENST00000362093.4 ENST00000417030.2 ENST00000527729.1 ENST00000532838.1 |
CKLF
CKLF-CMTM1
|
chemokine-like factor CKLF-CMTM1 readthrough |
| chr1_+_61547894 | 1.33 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr7_-_64023410 | 1.32 |
ENST00000447137.2
|
ZNF680
|
zinc finger protein 680 |
| chr13_+_37393351 | 1.29 |
ENST00000255476.2
|
RFXAP
|
regulatory factor X-associated protein |
| chr2_-_86116093 | 1.29 |
ENST00000377332.3
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr3_-_148804275 | 1.27 |
ENST00000392912.2
ENST00000465259.1 ENST00000310053.5 ENST00000494055.1 |
HLTF
|
helicase-like transcription factor |
| chrX_-_134429952 | 1.27 |
ENST00000370764.1
|
ZNF75D
|
zinc finger protein 75D |
| chr7_+_86975001 | 1.27 |
ENST00000412227.2
ENST00000331536.3 |
CROT
|
carnitine O-octanoyltransferase |
| chr2_+_113403434 | 1.25 |
ENST00000272542.3
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1 |
| chr2_+_170550944 | 1.25 |
ENST00000359744.3
ENST00000438838.1 ENST00000438710.1 ENST00000449906.1 ENST00000498202.2 ENST00000272797.4 |
PHOSPHO2
KLHL23
|
phosphatase, orphan 2 kelch-like family member 23 |
| chr3_-_15901278 | 1.25 |
ENST00000399451.2
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr1_+_65210772 | 1.24 |
ENST00000371072.4
ENST00000294428.3 |
RAVER2
|
ribonucleoprotein, PTB-binding 2 |
| chr2_+_163200598 | 1.24 |
ENST00000437150.2
ENST00000453113.2 |
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr8_-_29940628 | 1.24 |
ENST00000545648.1
ENST00000256255.6 |
TMEM66
|
transmembrane protein 66 |
| chr11_+_108093559 | 1.24 |
ENST00000278616.4
|
ATM
|
ataxia telangiectasia mutated |
| chr15_-_57210769 | 1.23 |
ENST00000559000.1
|
ZNF280D
|
zinc finger protein 280D |
| chrX_-_63005405 | 1.22 |
ENST00000374878.1
ENST00000437457.2 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr2_-_85829811 | 1.22 |
ENST00000306353.3
|
TMEM150A
|
transmembrane protein 150A |
| chr6_-_27440460 | 1.22 |
ENST00000377419.1
|
ZNF184
|
zinc finger protein 184 |
| chr2_-_170430277 | 1.21 |
ENST00000438035.1
ENST00000453929.2 |
FASTKD1
|
FAST kinase domains 1 |
| chr3_+_111697843 | 1.21 |
ENST00000534857.1
ENST00000273359.3 ENST00000494817.1 |
ABHD10
|
abhydrolase domain containing 10 |
| chr4_-_123843597 | 1.21 |
ENST00000510735.1
ENST00000304430.5 |
NUDT6
|
nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
| chr7_-_16685422 | 1.20 |
ENST00000306999.2
|
ANKMY2
|
ankyrin repeat and MYND domain containing 2 |
| chr6_+_30851840 | 1.20 |
ENST00000511510.1
ENST00000376569.3 ENST00000376575.3 ENST00000376570.4 ENST00000446312.1 ENST00000504927.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr7_+_17338239 | 1.20 |
ENST00000242057.4
|
AHR
|
aryl hydrocarbon receptor |
| chr11_+_46316677 | 1.19 |
ENST00000534787.1
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1 |
| chr14_+_61447832 | 1.19 |
ENST00000354886.2
ENST00000267488.4 |
SLC38A6
|
solute carrier family 38, member 6 |
| chr20_+_34700333 | 1.17 |
ENST00000441639.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr19_+_34745442 | 1.17 |
ENST00000299505.6
ENST00000588470.1 ENST00000589583.1 ENST00000588338.2 |
KIAA0355
|
KIAA0355 |
| chr11_-_105948040 | 1.15 |
ENST00000534815.1
|
KBTBD3
|
kelch repeat and BTB (POZ) domain containing 3 |
| chr1_-_25573937 | 1.14 |
ENST00000417642.2
ENST00000431849.2 |
C1orf63
|
chromosome 1 open reading frame 63 |
| chr3_-_194991876 | 1.14 |
ENST00000310380.6
|
XXYLT1
|
xyloside xylosyltransferase 1 |
| chr15_-_34659349 | 1.14 |
ENST00000314891.6
|
LPCAT4
|
lysophosphatidylcholine acyltransferase 4 |
| chr12_-_104531785 | 1.13 |
ENST00000551727.1
|
NFYB
|
nuclear transcription factor Y, beta |
| chr2_+_163200848 | 1.13 |
ENST00000233612.4
|
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr2_-_197036289 | 1.13 |
ENST00000263955.4
|
STK17B
|
serine/threonine kinase 17b |
| chr1_-_33647267 | 1.12 |
ENST00000291416.5
|
TRIM62
|
tripartite motif containing 62 |
| chr2_+_11295624 | 1.12 |
ENST00000402361.1
ENST00000428481.1 |
PQLC3
|
PQ loop repeat containing 3 |
| chr14_-_100772767 | 1.11 |
ENST00000392908.3
ENST00000539621.1 |
SLC25A29
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
| chr19_-_38146289 | 1.11 |
ENST00000392144.1
ENST00000591444.1 ENST00000351218.2 ENST00000587809.1 |
ZFP30
|
ZFP30 zinc finger protein |
| chr13_-_52585547 | 1.11 |
ENST00000448424.2
ENST00000400370.3 ENST00000418097.2 ENST00000242839.4 ENST00000400366.3 ENST00000344297.5 |
ATP7B
|
ATPase, Cu++ transporting, beta polypeptide |
| chr19_-_14201776 | 1.10 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr19_-_19051993 | 1.10 |
ENST00000594794.1
ENST00000355887.6 ENST00000392351.3 ENST00000596482.1 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr15_-_49338624 | 1.10 |
ENST00000261847.3
ENST00000380927.2 ENST00000559424.1 |
SECISBP2L
|
SECIS binding protein 2-like |
| chr4_+_9783252 | 1.09 |
ENST00000304374.2
|
DRD5
|
dopamine receptor D5 |
| chr9_+_139553306 | 1.09 |
ENST00000371699.1
|
EGFL7
|
EGF-like-domain, multiple 7 |
| chr1_-_209979465 | 1.09 |
ENST00000542854.1
|
IRF6
|
interferon regulatory factor 6 |
| chrX_-_57147748 | 1.09 |
ENST00000374910.3
|
SPIN2B
|
spindlin family, member 2B |
| chr5_-_107006596 | 1.08 |
ENST00000333274.6
|
EFNA5
|
ephrin-A5 |
| chr1_-_20812690 | 1.08 |
ENST00000375078.3
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr14_-_100772862 | 1.07 |
ENST00000359232.3
|
SLC25A29
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
| chr19_+_46001697 | 1.07 |
ENST00000451287.2
ENST00000324688.4 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr12_+_109826524 | 1.07 |
ENST00000431443.2
|
MYO1H
|
myosin IH |
| chr14_+_61447927 | 1.06 |
ENST00000451406.1
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr1_-_54411240 | 1.05 |
ENST00000371378.2
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr18_+_54318566 | 1.05 |
ENST00000589935.1
ENST00000357574.3 |
WDR7
|
WD repeat domain 7 |
| chr11_+_94822968 | 1.05 |
ENST00000278505.4
|
ENDOD1
|
endonuclease domain containing 1 |
| chr9_+_43684902 | 1.05 |
ENST00000377564.3
ENST00000276974.6 |
CNTNAP3B
|
contactin associated protein-like 3B |
| chr9_-_125693757 | 1.04 |
ENST00000373656.3
|
ZBTB26
|
zinc finger and BTB domain containing 26 |
| chr2_+_181845843 | 1.02 |
ENST00000602710.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr13_+_100741269 | 1.02 |
ENST00000376286.4
ENST00000376279.3 ENST00000376285.1 |
PCCA
|
propionyl CoA carboxylase, alpha polypeptide |
| chr13_+_50656307 | 1.01 |
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr11_-_69490135 | 1.01 |
ENST00000542341.1
|
ORAOV1
|
oral cancer overexpressed 1 |
| chr10_-_33623826 | 1.01 |
ENST00000374867.2
|
NRP1
|
neuropilin 1 |
| chr16_+_53088885 | 1.00 |
ENST00000566029.1
ENST00000447540.1 |
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr19_-_10491130 | 1.00 |
ENST00000530829.1
ENST00000529370.1 |
TYK2
|
tyrosine kinase 2 |
| chr4_-_169931393 | 1.00 |
ENST00000504480.1
ENST00000306193.3 |
CBR4
|
carbonyl reductase 4 |
| chr17_-_5487277 | 1.00 |
ENST00000572272.1
ENST00000354411.3 ENST00000577119.1 |
NLRP1
|
NLR family, pyrin domain containing 1 |
| chr8_-_127570603 | 1.00 |
ENST00000304916.3
|
FAM84B
|
family with sequence similarity 84, member B |
| chr12_+_82752275 | 0.99 |
ENST00000248306.3
|
METTL25
|
methyltransferase like 25 |
| chr6_+_160183492 | 0.98 |
ENST00000541436.1
|
ACAT2
|
acetyl-CoA acetyltransferase 2 |
| chr17_-_48943706 | 0.98 |
ENST00000499247.2
|
TOB1
|
transducer of ERBB2, 1 |
| chrX_-_16730688 | 0.98 |
ENST00000359276.4
|
CTPS2
|
CTP synthase 2 |
| chr1_+_162351503 | 0.98 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr5_-_43043272 | 0.98 |
ENST00000314890.3
|
ANXA2R
|
annexin A2 receptor |
| chr7_+_12250886 | 0.98 |
ENST00000444443.1
ENST00000396667.3 |
TMEM106B
|
transmembrane protein 106B |
| chr8_+_95835438 | 0.98 |
ENST00000521860.1
ENST00000519457.1 ENST00000519053.1 ENST00000523731.1 ENST00000447247.1 |
INTS8
|
integrator complex subunit 8 |
| chr2_-_209119831 | 0.97 |
ENST00000345146.2
|
IDH1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr3_+_44903361 | 0.97 |
ENST00000302392.4
|
TMEM42
|
transmembrane protein 42 |
| chr8_-_17941575 | 0.96 |
ENST00000417108.2
|
ASAH1
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1 |
| chr16_+_29827832 | 0.96 |
ENST00000609618.1
|
PAGR1
|
PAXIP1-associated glutamate-rich protein 1 |
| chr17_-_79869228 | 0.96 |
ENST00000570388.1
|
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr9_-_35812236 | 0.96 |
ENST00000340291.2
|
SPAG8
|
sperm associated antigen 8 |
| chr9_+_115142217 | 0.96 |
ENST00000398805.3
ENST00000398803.1 ENST00000262542.7 ENST00000539114.1 |
HSDL2
|
hydroxysteroid dehydrogenase like 2 |
| chr4_+_39460689 | 0.96 |
ENST00000381846.1
|
LIAS
|
lipoic acid synthetase |
| chr7_+_102715315 | 0.96 |
ENST00000428183.2
ENST00000323716.3 ENST00000441711.2 ENST00000454559.1 ENST00000425331.1 ENST00000541300.1 |
ARMC10
|
armadillo repeat containing 10 |
| chr7_+_65958693 | 0.96 |
ENST00000445681.1
ENST00000452565.1 |
GS1-124K5.4
|
GS1-124K5.4 |
| chr14_+_45605157 | 0.95 |
ENST00000542564.2
|
FANCM
|
Fanconi anemia, complementation group M |
| chr13_-_22178284 | 0.95 |
ENST00000468222.2
ENST00000382374.4 |
MICU2
|
mitochondrial calcium uptake 2 |
| chr15_-_31283798 | 0.95 |
ENST00000435680.1
ENST00000425768.1 |
MTMR10
|
myotubularin related protein 10 |
| chr17_+_73521763 | 0.95 |
ENST00000167462.5
ENST00000375227.4 ENST00000392550.3 ENST00000578363.1 ENST00000579392.1 |
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr7_-_143599207 | 0.95 |
ENST00000355951.2
ENST00000479870.1 ENST00000478172.1 |
FAM115A
|
family with sequence similarity 115, member A |
| chr5_+_53813536 | 0.94 |
ENST00000343017.6
ENST00000381410.4 ENST00000326277.3 |
SNX18
|
sorting nexin 18 |
| chr11_+_33563821 | 0.94 |
ENST00000321505.4
ENST00000265654.5 ENST00000389726.3 |
KIAA1549L
|
KIAA1549-like |
| chr2_-_132559234 | 0.94 |
ENST00000303798.2
|
C2orf27B
|
chromosome 2 open reading frame 27B |
| chr10_-_38146510 | 0.94 |
ENST00000395867.3
|
ZNF248
|
zinc finger protein 248 |
| chr5_-_138534071 | 0.94 |
ENST00000394817.2
|
SIL1
|
SIL1 nucleotide exchange factor |
| chr20_+_34042962 | 0.93 |
ENST00000446710.1
ENST00000420564.1 |
CEP250
|
centrosomal protein 250kDa |
| chr12_+_12870055 | 0.93 |
ENST00000228872.4
|
CDKN1B
|
cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
| chr5_+_64920826 | 0.93 |
ENST00000438419.2
ENST00000231526.4 ENST00000505553.1 ENST00000545191.1 |
TRAPPC13
|
trafficking protein particle complex 13 |
| chr17_+_47865917 | 0.93 |
ENST00000259021.4
ENST00000454930.2 ENST00000509773.1 ENST00000510819.1 ENST00000424009.2 |
KAT7
|
K(lysine) acetyltransferase 7 |
| chr8_-_28747717 | 0.92 |
ENST00000416984.2
|
INTS9
|
integrator complex subunit 9 |
| chr8_-_28747424 | 0.92 |
ENST00000523436.1
ENST00000397363.4 ENST00000521777.1 ENST00000520184.1 ENST00000521022.1 |
INTS9
|
integrator complex subunit 9 |
| chr11_-_17229480 | 0.92 |
ENST00000532035.1
ENST00000540361.1 |
PIK3C2A
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr1_-_54411255 | 0.92 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr17_-_79869004 | 0.91 |
ENST00000573927.1
ENST00000331285.3 ENST00000572157.1 |
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr1_-_231175964 | 0.91 |
ENST00000366654.4
|
FAM89A
|
family with sequence similarity 89, member A |
| chr2_-_69664549 | 0.91 |
ENST00000450796.2
ENST00000484177.1 |
NFU1
|
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr10_-_126849068 | 0.91 |
ENST00000494626.2
ENST00000337195.5 |
CTBP2
|
C-terminal binding protein 2 |
| chr3_+_23986748 | 0.91 |
ENST00000312521.4
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2F3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 10.2 | GO:2000690 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 1.4 | 11.5 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 1.3 | 4.0 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 1.2 | 4.8 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.9 | 2.6 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.8 | 2.5 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.7 | 2.2 | GO:0006844 | acyl carnitine transport(GO:0006844) histidine transport(GO:0015817) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) acyl carnitine transmembrane transport(GO:1902616) |
| 0.7 | 3.3 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.6 | 6.1 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.6 | 0.6 | GO:0003169 | coronary vein morphogenesis(GO:0003169) |
| 0.6 | 3.5 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.6 | 1.7 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.6 | 1.7 | GO:0071810 | circadian temperature homeostasis(GO:0060086) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.6 | 1.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.5 | 1.5 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.5 | 1.4 | GO:0090191 | negative regulation of mesonephros development(GO:0061218) negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.5 | 2.3 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.5 | 2.7 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.4 | 2.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.4 | 3.8 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.4 | 2.8 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.4 | 2.4 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.4 | 1.6 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.4 | 1.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.4 | 1.1 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.4 | 1.4 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.4 | 2.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.4 | 1.1 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.3 | 1.3 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.3 | 1.7 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.3 | 1.3 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.3 | 1.0 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.3 | 1.0 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.3 | 1.0 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.3 | 1.3 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.3 | 1.5 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.3 | 1.5 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.3 | 4.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.3 | 1.5 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.3 | 1.4 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.3 | 1.4 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.3 | 0.9 | GO:0002582 | myeloid dendritic cell activation involved in immune response(GO:0002277) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.3 | 0.9 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.3 | 3.4 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.3 | 0.8 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.3 | 1.7 | GO:0061441 | renal artery morphogenesis(GO:0061441) |
| 0.3 | 3.3 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.3 | 0.8 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.3 | 0.8 | GO:0046081 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.3 | 2.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.3 | 0.8 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.3 | 2.5 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.3 | 1.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.3 | 0.8 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 | 1.0 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.2 | 0.7 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.2 | 1.5 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.2 | 0.7 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.2 | 0.7 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.2 | 1.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.2 | 1.1 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.2 | 0.9 | GO:0061528 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.2 | 0.9 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.2 | 0.9 | GO:1905123 | regulation of endosome organization(GO:1904978) regulation of glucosylceramidase activity(GO:1905123) |
| 0.2 | 0.7 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.2 | 0.4 | GO:0061289 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.2 | 0.4 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.2 | 2.0 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 1.5 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.2 | 0.8 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.2 | 0.2 | GO:1904871 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.2 | 0.8 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.2 | 0.8 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 | 1.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.2 | 0.8 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.2 | 1.6 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 1.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.2 | 0.4 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.2 | 1.0 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.2 | 5.5 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 0.6 | GO:0019364 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.2 | 0.8 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.2 | 0.4 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.2 | 0.7 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.2 | 1.5 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.2 | 2.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.2 | 0.9 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.2 | 0.5 | GO:1903978 | regulation of microglial cell activation(GO:1903978) |
| 0.2 | 0.9 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.2 | 2.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.2 | 2.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.2 | 0.9 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 1.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 0.9 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 1.4 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.2 | 0.8 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.2 | 0.5 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.2 | 0.5 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.2 | 0.7 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.2 | 0.5 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.2 | 0.5 | GO:0046709 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.2 | 0.8 | GO:0019075 | virus maturation(GO:0019075) regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.2 | 0.6 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.2 | 0.5 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.2 | 0.5 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) maintenance of ER location(GO:0051685) |
| 0.2 | 0.9 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.2 | 1.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.2 | 2.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 0.5 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.6 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 0.7 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.4 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.1 | 0.3 | GO:0015911 | plasma membrane long-chain fatty acid transport(GO:0015911) |
| 0.1 | 0.3 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 1.7 | GO:0030647 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.1 | 3.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.7 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 1.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 0.3 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.4 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.1 | 0.8 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 2.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.5 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 1.7 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 1.6 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 1.3 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.1 | 0.6 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.1 | 0.9 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 1.3 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 0.5 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.6 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 1.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 1.9 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.5 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.5 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.7 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.9 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.5 | GO:0072396 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.1 | 1.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 | 0.3 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.1 | 0.8 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 1.2 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.1 | 0.4 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 0.3 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 0.6 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 1.1 | GO:1905206 | positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.1 | 0.4 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.1 | 0.4 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.1 | 0.5 | GO:0016128 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 0.1 | 0.7 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 1.4 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 0.4 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 0.9 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.1 | 0.6 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 1.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.5 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 2.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.6 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.4 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.1 | 0.3 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 0.6 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 0.8 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.1 | 1.0 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.8 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 1.6 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 0.8 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.3 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.6 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.3 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.1 | 0.3 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.1 | 1.5 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.1 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.1 | 0.7 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.4 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.1 | 0.4 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.1 | 0.9 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.3 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.1 | 0.4 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.1 | 0.3 | GO:2001162 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.1 | 0.3 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.1 | 2.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.2 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.1 | 0.5 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 0.3 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 | 0.3 | GO:0002752 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.6 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.1 | 0.5 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.1 | 1.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 2.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.0 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.8 | GO:1902065 | response to L-glutamate(GO:1902065) |
| 0.1 | 0.2 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.1 | 0.6 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.8 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 0.4 | GO:0030047 | actin modification(GO:0030047) |
| 0.1 | 0.7 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.6 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 1.0 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 1.8 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.4 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.1 | 0.3 | GO:1902949 | positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.1 | 0.3 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.1 | 1.2 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) |
| 0.1 | 0.2 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 0.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.2 | GO:2001035 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.1 | 0.3 | GO:0009397 | folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 0.3 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.4 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 | 0.4 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.1 | 0.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.2 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 0.8 | GO:0019227 | neuronal action potential propagation(GO:0019227) positive regulation of axon regeneration(GO:0048680) action potential propagation(GO:0098870) |
| 0.1 | 0.3 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.1 | 1.7 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.4 | GO:1902188 | positive regulation of viral release from host cell(GO:1902188) |
| 0.1 | 0.9 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.1 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 1.0 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 1.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.3 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.2 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.3 | GO:0050668 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.1 | 0.3 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 0.7 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 1.7 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.3 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 | 1.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.4 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.6 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.1 | 0.2 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 0.2 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.1 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.1 | 0.2 | GO:0035912 | dorsal aorta development(GO:0035907) dorsal aorta morphogenesis(GO:0035912) |
| 0.1 | 1.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 1.0 | GO:0019374 | galactosylceramide metabolic process(GO:0006681) galactolipid metabolic process(GO:0019374) |
| 0.1 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.2 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.1 | 0.4 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.1 | 0.2 | GO:0006106 | fumarate metabolic process(GO:0006106) glycerol biosynthetic process(GO:0006114) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) alditol biosynthetic process(GO:0019401) |
| 0.1 | 1.5 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.1 | 0.7 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.1 | 0.9 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 1.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 2.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 0.4 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.1 | 0.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.2 | GO:1904760 | peptidyl-serine ADP-ribosylation(GO:0018312) myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.1 | 0.4 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 0.4 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.6 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.1 | 2.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 1.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.2 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 0.3 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 3.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.2 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.1 | 0.3 | GO:2000232 | regulation of ribosome biogenesis(GO:0090069) positive regulation of ribosome biogenesis(GO:0090070) regulation of rRNA processing(GO:2000232) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 1.9 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 0.2 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 1.2 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 | 0.6 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.7 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.1 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.1 | 0.8 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 | 0.3 | GO:1901911 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.5 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.1 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.2 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.5 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.1 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 | 0.3 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.5 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.8 | GO:1902001 | fatty acid transmembrane transport(GO:1902001) |
| 0.0 | 0.4 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 2.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 1.5 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.0 | 1.8 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.4 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.8 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.0 | GO:0071106 | coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.0 | 0.3 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.2 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 1.0 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 4.0 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) |
| 0.0 | 0.9 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.1 | GO:1903939 | regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.2 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.3 | GO:1904180 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 1.0 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 3.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.4 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.5 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 0.6 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.8 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.8 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.0 | 0.1 | GO:0097195 | pilomotor reflex(GO:0097195) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.4 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.0 | 0.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 4.1 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.9 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.3 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.0 | 0.1 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.6 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.3 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.3 | GO:0010635 | regulation of mitochondrial fusion(GO:0010635) |
| 0.0 | 0.5 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.1 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.5 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.4 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 1.2 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.2 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.8 | GO:0036119 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.6 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.3 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) peptidyl-arginine omega-N-methylation(GO:0035247) |
| 0.0 | 0.4 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.2 | GO:0061741 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) |
| 0.0 | 0.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.3 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.3 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0035709 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 1.1 | GO:0046596 | regulation of viral entry into host cell(GO:0046596) |
| 0.0 | 0.2 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.7 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 1.4 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.2 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.4 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.0 | 0.1 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 1.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.4 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 0.0 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.0 | 0.3 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.3 | GO:0006555 | methionine metabolic process(GO:0006555) |
| 0.0 | 0.4 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.1 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.5 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) regulation of collagen catabolic process(GO:0010710) |
| 0.0 | 1.1 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.3 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.6 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.5 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 1.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.2 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.9 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.8 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:0090241 | negative regulation of histone H4 acetylation(GO:0090241) |
| 0.0 | 0.1 | GO:0044861 | protein localization to plasma membrane raft(GO:0044860) protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.0 | 0.3 | GO:0021930 | cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.0 | 0.2 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.0 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.3 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.4 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 0.0 | GO:0032899 | regulation of neurotrophin production(GO:0032899) |
| 0.0 | 0.1 | GO:0072537 | fibroblast activation(GO:0072537) |
| 0.0 | 0.1 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.0 | 0.7 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.0 | 0.1 | GO:1904806 | regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
| 0.0 | 0.5 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.3 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.4 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.3 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.7 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 1.0 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.0 | 0.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 1.4 | GO:0051297 | centrosome cycle(GO:0007098) centrosome organization(GO:0051297) |
| 0.0 | 0.1 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.0 | 0.6 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.0 | 1.0 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.6 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.2 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.2 | GO:0032462 | regulation of protein homooligomerization(GO:0032462) |
| 0.0 | 0.4 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.3 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.5 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.5 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 1.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.4 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.0 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.8 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.3 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.0 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.1 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 1.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.4 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.1 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:1904948 | midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.0 | 0.1 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.0 | 0.2 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.0 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.2 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.1 | GO:0033866 | nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.5 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.0 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.0 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.1 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0010919 | regulation of inositol phosphate biosynthetic process(GO:0010919) positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.0 | GO:0052330 | induction of programmed cell death(GO:0012502) peptidyl-cysteine S-trans-nitrosylation(GO:0035606) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.0 | 0.1 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.0 | 0.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.7 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 2.0 | GO:0008037 | cell recognition(GO:0008037) |
| 0.0 | 0.1 | GO:0051414 | luteolysis(GO:0001554) chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) response to cortisol(GO:0051414) apoptotic process involved in luteolysis(GO:0061364) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.5 | 1.4 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.4 | 1.7 | GO:0043293 | apoptosome(GO:0043293) |
| 0.4 | 1.7 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.4 | 1.5 | GO:0031251 | PAN complex(GO:0031251) |
| 0.3 | 0.6 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.3 | 1.9 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.3 | 1.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.3 | 0.8 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.3 | 1.6 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 1.3 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.3 | 3.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.3 | 11.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 1.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 2.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 0.7 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.2 | 0.9 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.2 | 1.5 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.2 | 2.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.2 | 0.8 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 0.6 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.2 | 1.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 2.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 2.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 3.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 0.9 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.2 | 0.7 | GO:0031213 | RSF complex(GO:0031213) |
| 0.2 | 0.6 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 0.5 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.2 | 2.0 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.2 | 0.5 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.2 | 0.5 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.2 | 0.5 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 4.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.8 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 1.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.7 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 1.9 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.4 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.1 | 0.7 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 2.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 2.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.4 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 0.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.4 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 1.6 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 1.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.6 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.8 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.7 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 1.0 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.1 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.1 | 3.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 2.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 1.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.4 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.3 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.1 | 0.6 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.5 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.3 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.9 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 0.3 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.4 | GO:0032449 | CBM complex(GO:0032449) |
| 0.1 | 1.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 5.2 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 1.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.2 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 1.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 1.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.4 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.4 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.2 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.3 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.1 | 4.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.4 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 7.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.3 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.5 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.2 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.1 | 0.2 | GO:1990742 | microvesicle(GO:1990742) |
| 0.1 | 0.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.2 | GO:0097196 | Shu complex(GO:0097196) |
| 0.1 | 0.4 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 1.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.4 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 3.8 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 3.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 3.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 4.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.6 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.4 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.2 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 1.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.0 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.6 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.3 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.6 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.9 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.8 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.0 | 0.3 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 2.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 1.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 2.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.5 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.2 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0019034 | viral replication complex(GO:0019034) host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.0 | 3.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.8 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.2 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 3.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.2 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.1 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 1.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.5 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.6 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 2.1 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 1.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.8 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.4 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.8 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.4 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.0 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 11.8 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 1.2 | 3.5 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 1.0 | 4.0 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.9 | 2.6 | GO:0016992 | lipoate synthase activity(GO:0016992) radical SAM enzyme activity(GO:0070283) |
| 0.7 | 2.2 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.7 | 3.4 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.6 | 3.2 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.6 | 1.8 | GO:0047025 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.5 | 1.6 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.5 | 2.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.5 | 1.6 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.4 | 2.5 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.4 | 1.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.4 | 1.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.3 | 0.7 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.3 | 1.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.3 | 1.0 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.3 | 1.0 | GO:0051990 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.3 | 2.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 2.5 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.3 | 2.5 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.3 | 1.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.3 | 1.5 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.3 | 1.8 | GO:0004096 | catalase activity(GO:0004096) |
| 0.3 | 0.9 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.3 | 0.9 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.3 | 2.2 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.3 | 1.4 | GO:0032142 | single guanine insertion binding(GO:0032142) |
| 0.3 | 0.8 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.3 | 3.0 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.3 | 0.8 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.3 | 0.8 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.3 | 1.0 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.3 | 1.8 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.3 | 1.0 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.2 | 0.7 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 1.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.2 | 0.7 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.2 | 1.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.2 | 0.7 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) leukemia inhibitory factor receptor activity(GO:0004923) interleukin-6 binding(GO:0019981) |
| 0.2 | 0.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 0.2 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.2 | 0.9 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.2 | 0.9 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.2 | 0.9 | GO:0004057 | arginyltransferase activity(GO:0004057) |
| 0.2 | 0.7 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.2 | 0.6 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.2 | 0.8 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.2 | 0.6 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.2 | 0.8 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.2 | 0.2 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.2 | 0.6 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.2 | 1.9 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.2 | 1.0 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.2 | 1.4 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.2 | 1.0 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.2 | 0.8 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.2 | 1.0 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 1.3 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.2 | 5.4 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.2 | 0.6 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.2 | 2.9 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 0.7 | GO:0004566 | beta-glucuronidase activity(GO:0004566) heparanase activity(GO:0030305) |
| 0.2 | 2.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 2.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 0.5 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.2 | 0.5 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 0.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 1.6 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.2 | 0.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 0.6 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 1.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.9 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.4 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 0.7 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 6.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.8 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 1.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.4 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.8 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.4 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.8 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.4 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.8 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.9 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 1.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.4 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.1 | 0.5 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.1 | 0.5 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.1 | 0.4 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.1 | 0.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 2.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.5 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 1.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.4 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.1 | 0.5 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.5 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 0.6 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 1.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.4 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.1 | 0.6 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.3 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 0.5 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.6 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 1.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.3 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.1 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.4 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 2.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.4 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.1 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.4 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.1 | 0.3 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.1 | 1.3 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.4 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.4 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 0.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.4 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.4 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.4 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.3 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.1 | 0.3 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.1 | 0.3 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 1.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.1 | 0.9 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 0.2 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.1 | 0.2 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.1 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.3 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.3 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.1 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 1.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.5 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.7 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 1.0 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 1.0 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.2 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 1.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.4 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 1.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.8 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.4 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 0.4 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 2.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.2 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.1 | 0.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.2 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 2.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.9 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.5 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) phosphatidylserine decarboxylase activity(GO:0004609) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.2 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.1 | 1.3 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 0.4 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.8 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.2 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.4 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.3 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 1.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 0.2 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 1.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 3.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.3 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.5 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.1 | 0.3 | GO:0052845 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.1 | 0.3 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 1.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.5 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.4 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 1.0 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.5 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.6 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.4 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.3 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.5 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.1 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.5 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.3 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.2 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 1.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 3.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 1.0 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.7 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 0.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 2.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.5 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.5 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.7 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 2.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.2 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 4.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0036317 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.2 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 2.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.3 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.1 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.2 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.9 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.5 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.1 | GO:0016855 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 1.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0005503 | all-trans retinal binding(GO:0005503) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 2.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0052830 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 1.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 1.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 2.1 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.8 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.4 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.3 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.7 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.3 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.0 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.0 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.0 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.0 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 1.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 1.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 4.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 7.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 0.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 3.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 1.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.3 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 2.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 3.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 2.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 2.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.7 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.2 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 1.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 2.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 3.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 3.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.2 | 3.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 5.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 3.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 4.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.6 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 4.6 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 2.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.9 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.8 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 1.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.7 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 1.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 1.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 5.5 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.1 | 2.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.8 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 3.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 0.5 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.1 | 0.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 4.5 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 1.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.9 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 1.0 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 1.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.7 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.9 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.0 | 1.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 1.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.7 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 2.0 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.8 | REACTOME GLOBAL GENOMIC NER GG NER | Genes involved in Global Genomic NER (GG-NER) |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 8.6 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.5 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 1.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR MUTANTS | Genes involved in Signaling by FGFR mutants |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |