Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for E2F4
Z-value: 0.75
Transcription factors associated with E2F4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2F4
|
ENSG00000205250.4 | E2F transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2F4 | hg19_v2_chr16_+_67226019_67226127 | 0.17 | 4.2e-01 | Click! |
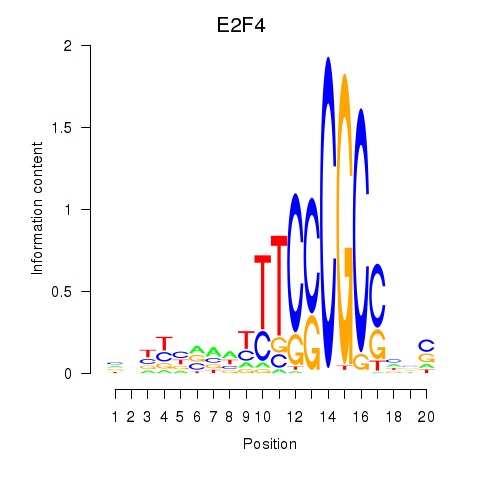
Activity profile of E2F4 motif
Sorted Z-values of E2F4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_203736150 | 1.79 |
ENST00000457524.1
ENST00000421334.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr4_-_157892167 | 1.15 |
ENST00000541126.1
|
PDGFC
|
platelet derived growth factor C |
| chr19_+_18284477 | 1.06 |
ENST00000407280.3
|
IFI30
|
interferon, gamma-inducible protein 30 |
| chr12_-_57472522 | 1.04 |
ENST00000379391.3
ENST00000300128.4 |
TMEM194A
|
transmembrane protein 194A |
| chr4_-_157892498 | 0.98 |
ENST00000502773.1
|
PDGFC
|
platelet derived growth factor C |
| chr2_-_133427767 | 0.95 |
ENST00000397463.2
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr9_+_35732312 | 0.94 |
ENST00000353704.2
|
CREB3
|
cAMP responsive element binding protein 3 |
| chr14_+_21538517 | 0.91 |
ENST00000298693.3
|
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr10_-_62493223 | 0.90 |
ENST00000373827.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr4_-_157892055 | 0.90 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr12_-_77459306 | 0.87 |
ENST00000547316.1
ENST00000416496.2 ENST00000550669.1 ENST00000322886.7 |
E2F7
|
E2F transcription factor 7 |
| chr19_-_6424783 | 0.83 |
ENST00000398148.3
|
KHSRP
|
KH-type splicing regulatory protein |
| chr14_+_21538429 | 0.76 |
ENST00000298694.4
ENST00000555038.1 |
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr7_-_108096765 | 0.74 |
ENST00000379024.4
ENST00000351718.4 |
NRCAM
|
neuronal cell adhesion molecule |
| chr11_+_82612740 | 0.72 |
ENST00000524921.1
ENST00000528759.1 ENST00000525361.1 ENST00000430323.2 ENST00000533655.1 ENST00000532764.1 ENST00000532589.1 ENST00000525388.1 |
C11orf82
|
chromosome 11 open reading frame 82 |
| chr7_-_108096822 | 0.69 |
ENST00000379028.3
ENST00000413765.2 ENST00000379022.4 |
NRCAM
|
neuronal cell adhesion molecule |
| chr10_+_122216316 | 0.66 |
ENST00000398250.1
ENST00000439221.1 ENST00000398248.1 |
PPAPDC1A
|
phosphatidic acid phosphatase type 2 domain containing 1A |
| chr17_+_43238438 | 0.66 |
ENST00000593138.1
ENST00000586681.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr4_+_74702214 | 0.63 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr15_+_92937144 | 0.63 |
ENST00000539113.1
ENST00000555434.1 |
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr7_+_138145145 | 0.62 |
ENST00000415680.2
|
TRIM24
|
tripartite motif containing 24 |
| chr3_-_113415441 | 0.61 |
ENST00000491165.1
ENST00000316407.4 |
KIAA2018
|
KIAA2018 |
| chr9_-_21994344 | 0.60 |
ENST00000530628.2
ENST00000361570.3 |
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr8_-_119964434 | 0.60 |
ENST00000297350.4
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b |
| chr4_-_120549163 | 0.60 |
ENST00000394439.1
ENST00000420633.1 |
PDE5A
|
phosphodiesterase 5A, cGMP-specific |
| chr15_+_91260552 | 0.60 |
ENST00000355112.3
ENST00000560509.1 |
BLM
|
Bloom syndrome, RecQ helicase-like |
| chr1_-_197115818 | 0.59 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr8_+_124084899 | 0.54 |
ENST00000287380.1
ENST00000309336.3 ENST00000519418.1 ENST00000327098.5 ENST00000522420.1 ENST00000521676.1 ENST00000378080.2 |
TBC1D31
|
TBC1 domain family, member 31 |
| chr4_-_1714037 | 0.54 |
ENST00000488267.1
ENST00000429429.2 ENST00000480936.1 |
SLBP
|
stem-loop binding protein |
| chr14_-_55658323 | 0.53 |
ENST00000554067.1
ENST00000247191.2 |
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr7_+_138145076 | 0.53 |
ENST00000343526.4
|
TRIM24
|
tripartite motif containing 24 |
| chr11_-_34379546 | 0.53 |
ENST00000435224.2
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2 |
| chr1_+_113217345 | 0.53 |
ENST00000357443.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr4_-_2758015 | 0.53 |
ENST00000510267.1
ENST00000503235.1 ENST00000315423.7 |
TNIP2
|
TNFAIP3 interacting protein 2 |
| chr1_+_113217309 | 0.52 |
ENST00000544796.1
ENST00000369644.1 |
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr16_+_50776021 | 0.52 |
ENST00000566679.2
ENST00000564634.1 ENST00000398568.2 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr21_-_44496441 | 0.52 |
ENST00000359624.3
ENST00000352178.5 |
CBS
|
cystathionine-beta-synthase |
| chr5_+_148521381 | 0.52 |
ENST00000504238.1
|
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr6_-_137113604 | 0.51 |
ENST00000359015.4
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr2_+_154728426 | 0.50 |
ENST00000392825.3
ENST00000434213.1 |
GALNT13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 (GalNAc-T13) |
| chr12_+_49717019 | 0.49 |
ENST00000549275.1
ENST00000551245.1 ENST00000380327.5 ENST00000548311.1 ENST00000550346.1 ENST00000550709.1 ENST00000549534.1 ENST00000257909.3 |
TROAP
|
trophinin associated protein |
| chr6_+_45389893 | 0.49 |
ENST00000371432.3
|
RUNX2
|
runt-related transcription factor 2 |
| chr1_-_28241024 | 0.49 |
ENST00000313433.7
ENST00000444045.1 |
RPA2
|
replication protein A2, 32kDa |
| chr1_-_226496898 | 0.49 |
ENST00000481685.1
|
LIN9
|
lin-9 homolog (C. elegans) |
| chr11_+_58910295 | 0.48 |
ENST00000420244.1
|
FAM111A
|
family with sequence similarity 111, member A |
| chr19_-_10444188 | 0.48 |
ENST00000293677.6
|
RAVER1
|
ribonucleoprotein, PTB-binding 1 |
| chr1_-_229761717 | 0.48 |
ENST00000366675.3
ENST00000258281.2 ENST00000366674.1 |
TAF5L
|
TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa |
| chrX_+_30671476 | 0.47 |
ENST00000378946.3
ENST00000378943.3 ENST00000378945.3 ENST00000427190.1 ENST00000378941.3 |
GK
|
glycerol kinase |
| chr6_+_126661253 | 0.47 |
ENST00000368326.1
ENST00000368325.1 ENST00000368328.4 |
CENPW
|
centromere protein W |
| chr1_-_33283754 | 0.47 |
ENST00000373477.4
|
YARS
|
tyrosyl-tRNA synthetase |
| chr14_+_59104741 | 0.46 |
ENST00000395153.3
ENST00000335867.4 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr3_+_179280668 | 0.46 |
ENST00000429709.2
ENST00000450518.2 ENST00000392662.1 ENST00000490364.1 |
ACTL6A
|
actin-like 6A |
| chr17_+_45810594 | 0.46 |
ENST00000177694.1
|
TBX21
|
T-box 21 |
| chr1_+_184356188 | 0.46 |
ENST00000235307.6
|
C1orf21
|
chromosome 1 open reading frame 21 |
| chr9_-_35115836 | 0.46 |
ENST00000378566.1
ENST00000378554.2 ENST00000322813.5 |
FAM214B
|
family with sequence similarity 214, member B |
| chr12_+_49717081 | 0.45 |
ENST00000547807.1
ENST00000551567.1 |
TROAP
|
trophinin associated protein |
| chr4_-_185655278 | 0.45 |
ENST00000281453.5
|
MLF1IP
|
centromere protein U |
| chr2_+_47168313 | 0.44 |
ENST00000319190.5
ENST00000394850.2 ENST00000536057.1 |
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr16_+_50775971 | 0.44 |
ENST00000311559.9
ENST00000564326.1 ENST00000566206.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr6_-_27114577 | 0.44 |
ENST00000356950.1
ENST00000396891.4 |
HIST1H2BK
|
histone cluster 1, H2bk |
| chr16_-_67693846 | 0.43 |
ENST00000602850.1
|
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chr2_+_71357434 | 0.43 |
ENST00000244230.2
|
MPHOSPH10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr11_-_123525289 | 0.42 |
ENST00000392770.2
ENST00000299333.3 ENST00000530277.1 |
SCN3B
|
sodium channel, voltage-gated, type III, beta subunit |
| chr7_-_27135591 | 0.42 |
ENST00000343060.4
ENST00000355633.5 |
HOXA1
|
homeobox A1 |
| chr6_-_139308777 | 0.42 |
ENST00000529597.1
ENST00000415951.2 ENST00000367663.4 ENST00000409812.2 |
REPS1
|
RALBP1 associated Eps domain containing 1 |
| chr6_-_31632962 | 0.42 |
ENST00000456540.1
ENST00000445768.1 |
GPANK1
|
G patch domain and ankyrin repeats 1 |
| chr19_-_10679697 | 0.42 |
ENST00000335766.2
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr3_+_133292574 | 0.41 |
ENST00000264993.3
|
CDV3
|
CDV3 homolog (mouse) |
| chr19_+_58694396 | 0.41 |
ENST00000326804.4
ENST00000345813.3 ENST00000424679.2 |
ZNF274
|
zinc finger protein 274 |
| chr1_-_28241226 | 0.41 |
ENST00000373912.3
ENST00000373909.3 |
RPA2
|
replication protein A2, 32kDa |
| chr17_-_76183111 | 0.41 |
ENST00000405273.1
ENST00000590862.1 ENST00000590430.1 ENST00000586613.1 |
TK1
|
thymidine kinase 1, soluble |
| chr19_-_45909585 | 0.40 |
ENST00000593226.1
ENST00000418234.2 |
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like |
| chr1_+_179262905 | 0.40 |
ENST00000539888.1
ENST00000540564.1 ENST00000535686.1 ENST00000367619.3 |
SOAT1
|
sterol O-acyltransferase 1 |
| chr6_-_139309378 | 0.40 |
ENST00000450536.2
ENST00000258062.5 |
REPS1
|
RALBP1 associated Eps domain containing 1 |
| chr19_-_10679644 | 0.39 |
ENST00000393599.2
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr6_+_43739697 | 0.39 |
ENST00000230480.6
|
VEGFA
|
vascular endothelial growth factor A |
| chr15_+_79724858 | 0.39 |
ENST00000305428.3
|
KIAA1024
|
KIAA1024 |
| chr1_-_44497024 | 0.39 |
ENST00000372306.3
ENST00000372310.3 ENST00000475075.2 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr7_-_129845313 | 0.39 |
ENST00000397622.2
|
TMEM209
|
transmembrane protein 209 |
| chr4_-_1713977 | 0.38 |
ENST00000318386.4
|
SLBP
|
stem-loop binding protein |
| chr15_+_76196200 | 0.38 |
ENST00000308275.3
ENST00000453211.2 |
FBXO22
|
F-box protein 22 |
| chr15_+_90744533 | 0.37 |
ENST00000411539.2
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr19_+_50887585 | 0.37 |
ENST00000440232.2
ENST00000601098.1 ENST00000599857.1 ENST00000593887.1 |
POLD1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr8_-_132052458 | 0.36 |
ENST00000377928.3
|
ADCY8
|
adenylate cyclase 8 (brain) |
| chr6_+_83073952 | 0.36 |
ENST00000543496.1
|
TPBG
|
trophoblast glycoprotein |
| chr16_+_50775948 | 0.35 |
ENST00000569681.1
ENST00000569418.1 ENST00000540145.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr5_-_79950371 | 0.35 |
ENST00000511032.1
ENST00000504396.1 ENST00000505337.1 |
DHFR
|
dihydrofolate reductase |
| chr5_-_149682447 | 0.35 |
ENST00000328668.7
|
ARSI
|
arylsulfatase family, member I |
| chr4_+_166794862 | 0.35 |
ENST00000513213.1
|
TLL1
|
tolloid-like 1 |
| chr15_+_76196234 | 0.34 |
ENST00000540507.1
ENST00000565036.1 ENST00000569054.1 |
FBXO22
|
F-box protein 22 |
| chr15_+_36871806 | 0.34 |
ENST00000566621.1
ENST00000564586.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr7_+_73703728 | 0.34 |
ENST00000361545.5
ENST00000223398.6 |
CLIP2
|
CAP-GLY domain containing linker protein 2 |
| chr18_+_33767473 | 0.33 |
ENST00000261326.5
|
MOCOS
|
molybdenum cofactor sulfurase |
| chr16_-_67694129 | 0.33 |
ENST00000602320.1
|
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chr11_-_126870655 | 0.33 |
ENST00000525144.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr15_+_42841008 | 0.33 |
ENST00000260372.3
ENST00000568876.1 ENST00000568846.2 ENST00000562398.1 |
HAUS2
|
HAUS augmin-like complex, subunit 2 |
| chr13_-_41706864 | 0.33 |
ENST00000379485.1
ENST00000499385.2 |
KBTBD6
|
kelch repeat and BTB (POZ) domain containing 6 |
| chr21_-_45660723 | 0.33 |
ENST00000344330.4
ENST00000407780.3 ENST00000400379.3 |
ICOSLG
|
inducible T-cell co-stimulator ligand |
| chr12_-_121734489 | 0.33 |
ENST00000412367.2
ENST00000402834.4 ENST00000404169.3 |
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr9_+_504674 | 0.32 |
ENST00000382297.2
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr20_+_58508817 | 0.32 |
ENST00000358293.3
|
FAM217B
|
family with sequence similarity 217, member B |
| chr1_+_10490779 | 0.32 |
ENST00000477755.1
|
APITD1
|
apoptosis-inducing, TAF9-like domain 1 |
| chr9_-_35732362 | 0.32 |
ENST00000314888.9
ENST00000540444.1 |
TLN1
|
talin 1 |
| chr11_-_64851496 | 0.31 |
ENST00000404147.3
ENST00000275517.3 |
CDCA5
|
cell division cycle associated 5 |
| chr5_+_156887027 | 0.31 |
ENST00000435489.2
ENST00000311946.7 |
NIPAL4
|
NIPA-like domain containing 4 |
| chr12_-_53574671 | 0.31 |
ENST00000444623.1
|
CSAD
|
cysteine sulfinic acid decarboxylase |
| chr16_+_67193834 | 0.31 |
ENST00000258200.3
ENST00000518148.1 ENST00000519917.1 ENST00000517382.1 ENST00000521920.1 |
FBXL8
|
F-box and leucine-rich repeat protein 8 |
| chr1_+_10490441 | 0.31 |
ENST00000470413.2
ENST00000309048.3 |
APITD1-CORT
APITD1
|
APITD1-CORT readthrough apoptosis-inducing, TAF9-like domain 1 |
| chr15_-_50647370 | 0.31 |
ENST00000558970.1
ENST00000396464.3 ENST00000560825.1 |
GABPB1
|
GA binding protein transcription factor, beta subunit 1 |
| chr3_+_140770183 | 0.30 |
ENST00000310546.2
|
SPSB4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr9_-_113018746 | 0.30 |
ENST00000374515.5
|
TXN
|
thioredoxin |
| chr3_-_27525865 | 0.30 |
ENST00000445684.1
|
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr11_-_61735103 | 0.30 |
ENST00000529191.1
ENST00000529631.1 ENST00000530019.1 ENST00000529548.1 ENST00000273550.7 |
FTH1
|
ferritin, heavy polypeptide 1 |
| chr2_-_203736452 | 0.30 |
ENST00000419460.1
|
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr5_+_14664762 | 0.30 |
ENST00000284274.4
|
FAM105B
|
family with sequence similarity 105, member B |
| chrX_+_109245863 | 0.30 |
ENST00000372072.3
|
TMEM164
|
transmembrane protein 164 |
| chrX_-_110039038 | 0.29 |
ENST00000372042.1
ENST00000482160.1 ENST00000444321.2 ENST00000218054.4 |
CHRDL1
|
chordin-like 1 |
| chr1_+_27248203 | 0.29 |
ENST00000321265.5
|
NUDC
|
nudC nuclear distribution protein |
| chr15_+_71184931 | 0.29 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr13_+_32889605 | 0.29 |
ENST00000380152.3
ENST00000544455.1 ENST00000530893.2 |
BRCA2
|
breast cancer 2, early onset |
| chrX_-_110038990 | 0.29 |
ENST00000372045.1
ENST00000394797.4 |
CHRDL1
|
chordin-like 1 |
| chr8_+_128748466 | 0.28 |
ENST00000524013.1
ENST00000520751.1 |
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr1_-_156647189 | 0.28 |
ENST00000368223.3
|
NES
|
nestin |
| chrX_-_110039286 | 0.28 |
ENST00000434224.1
|
CHRDL1
|
chordin-like 1 |
| chr4_-_120548779 | 0.28 |
ENST00000264805.5
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific |
| chrX_-_40594755 | 0.28 |
ENST00000324817.1
|
MED14
|
mediator complex subunit 14 |
| chr15_+_36871983 | 0.28 |
ENST00000437989.2
ENST00000569302.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr19_-_8070474 | 0.27 |
ENST00000407627.2
ENST00000593807.1 |
ELAVL1
|
ELAV like RNA binding protein 1 |
| chr8_+_128748308 | 0.27 |
ENST00000377970.2
|
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr16_+_30064411 | 0.27 |
ENST00000338110.5
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr4_-_119757322 | 0.27 |
ENST00000379735.5
|
SEC24D
|
SEC24 family member D |
| chr15_-_50647274 | 0.27 |
ENST00000543881.1
|
GABPB1
|
GA binding protein transcription factor, beta subunit 1 |
| chr16_-_67694597 | 0.26 |
ENST00000393919.4
ENST00000219251.8 |
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chr1_-_204654481 | 0.26 |
ENST00000367176.3
|
LRRN2
|
leucine rich repeat neuronal 2 |
| chr16_+_30064444 | 0.26 |
ENST00000395248.1
ENST00000566897.1 ENST00000568435.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr11_+_32914579 | 0.26 |
ENST00000399302.2
|
QSER1
|
glutamine and serine rich 1 |
| chr1_+_109756523 | 0.26 |
ENST00000234677.2
ENST00000369923.4 |
SARS
|
seryl-tRNA synthetase |
| chr19_-_14016877 | 0.25 |
ENST00000454313.1
ENST00000591586.1 ENST00000346736.2 |
C19orf57
|
chromosome 19 open reading frame 57 |
| chr1_-_26185844 | 0.25 |
ENST00000538789.1
ENST00000374298.3 |
AUNIP
|
aurora kinase A and ninein interacting protein |
| chr15_+_71185148 | 0.25 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr1_+_11333245 | 0.25 |
ENST00000376810.5
|
UBIAD1
|
UbiA prenyltransferase domain containing 1 |
| chr20_+_47662805 | 0.25 |
ENST00000262982.2
ENST00000542325.1 |
CSE1L
|
CSE1 chromosome segregation 1-like (yeast) |
| chr3_-_27525826 | 0.24 |
ENST00000454389.1
ENST00000440156.1 ENST00000437179.1 ENST00000446700.1 ENST00000455077.1 ENST00000435667.2 ENST00000388777.4 ENST00000425128.2 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr7_-_149158187 | 0.24 |
ENST00000247930.4
|
ZNF777
|
zinc finger protein 777 |
| chr7_-_98741714 | 0.24 |
ENST00000361125.1
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr11_+_62623544 | 0.24 |
ENST00000377890.2
ENST00000377891.2 ENST00000377889.2 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr6_-_71666732 | 0.24 |
ENST00000230053.6
|
B3GAT2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr8_+_17104539 | 0.24 |
ENST00000521829.1
ENST00000521005.1 |
VPS37A
|
vacuolar protein sorting 37 homolog A (S. cerevisiae) |
| chr2_+_102803433 | 0.24 |
ENST00000264257.2
ENST00000441515.2 |
IL1RL2
|
interleukin 1 receptor-like 2 |
| chr9_-_136857403 | 0.24 |
ENST00000406606.3
ENST00000371850.3 |
VAV2
|
vav 2 guanine nucleotide exchange factor |
| chr6_+_45390222 | 0.24 |
ENST00000359524.5
|
RUNX2
|
runt-related transcription factor 2 |
| chr12_-_48499591 | 0.24 |
ENST00000551330.1
ENST00000004980.5 ENST00000339976.6 ENST00000448372.1 |
SENP1
|
SUMO1/sentrin specific peptidase 1 |
| chr16_+_11038403 | 0.23 |
ENST00000409552.3
|
CLEC16A
|
C-type lectin domain family 16, member A |
| chr19_+_17392672 | 0.23 |
ENST00000594072.1
ENST00000598347.1 |
ANKLE1
|
ankyrin repeat and LEM domain containing 1 |
| chr16_-_67194201 | 0.23 |
ENST00000345057.4
|
TRADD
|
TNFRSF1A-associated via death domain |
| chr9_-_21994597 | 0.23 |
ENST00000579755.1
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr3_+_10068095 | 0.23 |
ENST00000287647.3
ENST00000383807.1 ENST00000383806.1 ENST00000419585.1 |
FANCD2
|
Fanconi anemia, complementation group D2 |
| chr5_+_109025067 | 0.23 |
ENST00000261483.4
|
MAN2A1
|
mannosidase, alpha, class 2A, member 1 |
| chr3_-_122233723 | 0.23 |
ENST00000493510.1
ENST00000344337.6 ENST00000476916.1 ENST00000465882.1 |
KPNA1
|
karyopherin alpha 1 (importin alpha 5) |
| chr1_+_28844778 | 0.23 |
ENST00000411533.1
|
RCC1
|
regulator of chromosome condensation 1 |
| chrX_-_20159934 | 0.23 |
ENST00000379593.1
ENST00000379607.5 |
EIF1AX
|
eukaryotic translation initiation factor 1A, X-linked |
| chr3_-_169899504 | 0.23 |
ENST00000474275.1
ENST00000484931.1 ENST00000494943.1 ENST00000497658.1 ENST00000465896.1 ENST00000475729.1 ENST00000495893.2 ENST00000481639.1 ENST00000467570.1 ENST00000466189.1 |
PHC3
|
polyhomeotic homolog 3 (Drosophila) |
| chr16_-_4401258 | 0.23 |
ENST00000577031.1
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr11_+_62623512 | 0.23 |
ENST00000377892.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr11_+_85955787 | 0.22 |
ENST00000528180.1
|
EED
|
embryonic ectoderm development |
| chr11_+_62623621 | 0.22 |
ENST00000535296.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr18_-_658244 | 0.22 |
ENST00000585033.1
ENST00000323813.3 |
C18orf56
|
chromosome 18 open reading frame 56 |
| chr1_-_145076186 | 0.22 |
ENST00000369348.3
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr6_-_82957433 | 0.22 |
ENST00000306270.7
|
IBTK
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr1_-_10003372 | 0.22 |
ENST00000377223.1
ENST00000541052.1 ENST00000377213.1 |
LZIC
|
leucine zipper and CTNNBIP1 domain containing |
| chr20_-_39995467 | 0.21 |
ENST00000332312.3
|
EMILIN3
|
elastin microfibril interfacer 3 |
| chr10_-_88281494 | 0.21 |
ENST00000298767.5
|
WAPAL
|
wings apart-like homolog (Drosophila) |
| chr15_-_50647347 | 0.21 |
ENST00000220429.8
ENST00000429662.2 |
GABPB1
|
GA binding protein transcription factor, beta subunit 1 |
| chr16_+_699319 | 0.21 |
ENST00000549091.1
ENST00000293879.4 |
WDR90
|
WD repeat domain 90 |
| chrX_+_133594168 | 0.21 |
ENST00000298556.7
|
HPRT1
|
hypoxanthine phosphoribosyltransferase 1 |
| chr5_-_127873659 | 0.21 |
ENST00000262464.4
|
FBN2
|
fibrillin 2 |
| chr1_-_26372602 | 0.21 |
ENST00000374278.3
ENST00000374276.3 |
SLC30A2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr13_+_95254085 | 0.21 |
ENST00000376958.4
|
GPR180
|
G protein-coupled receptor 180 |
| chr4_-_73434498 | 0.20 |
ENST00000286657.4
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr19_+_16187085 | 0.20 |
ENST00000300933.4
|
TPM4
|
tropomyosin 4 |
| chr19_-_19754404 | 0.20 |
ENST00000587205.1
ENST00000445806.2 ENST00000203556.4 |
GMIP
|
GEM interacting protein |
| chr2_+_178077477 | 0.20 |
ENST00000411529.2
ENST00000435711.1 |
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr1_-_241683001 | 0.20 |
ENST00000366560.3
|
FH
|
fumarate hydratase |
| chr1_-_47779762 | 0.20 |
ENST00000371877.3
ENST00000360380.3 ENST00000337817.5 ENST00000447475.2 |
STIL
|
SCL/TAL1 interrupting locus |
| chr1_+_10490127 | 0.20 |
ENST00000602787.1
ENST00000602296.1 ENST00000400900.2 |
APITD1
APITD1-CORT
|
apoptosis-inducing, TAF9-like domain 1 APITD1-CORT readthrough |
| chr4_-_83821676 | 0.20 |
ENST00000355196.2
ENST00000507676.1 ENST00000506495.1 ENST00000507051.1 |
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr12_-_125052010 | 0.20 |
ENST00000458234.1
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr10_-_70166946 | 0.20 |
ENST00000388768.2
|
RUFY2
|
RUN and FYVE domain containing 2 |
| chr1_-_204654826 | 0.20 |
ENST00000367177.3
|
LRRN2
|
leucine rich repeat neuronal 2 |
| chr4_+_39460689 | 0.19 |
ENST00000381846.1
|
LIAS
|
lipoic acid synthetase |
| chr3_+_52719936 | 0.19 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr7_-_102985288 | 0.19 |
ENST00000379263.3
|
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr8_+_145743360 | 0.19 |
ENST00000527730.1
ENST00000529022.1 ENST00000292524.1 |
LRRC14
|
leucine rich repeat containing 14 |
| chr19_+_797443 | 0.19 |
ENST00000394601.4
ENST00000589575.1 |
PTBP1
|
polypyrimidine tract binding protein 1 |
| chr3_-_100120223 | 0.19 |
ENST00000284320.5
|
TOMM70A
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) |
| chr1_+_151254738 | 0.19 |
ENST00000336715.6
ENST00000324048.5 ENST00000368879.2 |
ZNF687
|
zinc finger protein 687 |
| chr7_-_99698338 | 0.19 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr5_+_141348721 | 0.19 |
ENST00000507163.1
ENST00000394519.1 |
RNF14
|
ring finger protein 14 |
| chr17_+_73008755 | 0.19 |
ENST00000584208.1
ENST00000301585.5 |
ICT1
|
immature colon carcinoma transcript 1 |
| chr11_+_85956182 | 0.19 |
ENST00000327320.4
ENST00000351625.6 ENST00000534595.1 |
EED
|
embryonic ectoderm development |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2F4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.3 | 1.0 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.3 | 0.9 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.2 | 0.9 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 0.6 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.2 | 0.6 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.2 | 0.9 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.2 | 0.6 | GO:0072186 | metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) |
| 0.2 | 0.5 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.2 | 0.9 | GO:0048749 | compound eye development(GO:0048749) |
| 0.2 | 0.5 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.2 | 1.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.2 | 0.3 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.2 | 0.5 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.1 | 0.7 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 0.4 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.4 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.1 | 1.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.9 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.9 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.3 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.1 | 0.7 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 0.1 | 3.0 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.3 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.4 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.4 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 1.1 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.3 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.5 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 0.5 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 0.3 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.1 | 0.3 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 | 0.8 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.1 | 0.8 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.1 | 0.7 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.4 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.8 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.1 | 0.2 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 0.8 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.1 | 0.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.2 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.1 | 0.3 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.1 | 0.4 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 0.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.5 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.2 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 0.2 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.1 | 0.2 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 | 0.2 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.2 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.5 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.4 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.0 | 0.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.3 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.0 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.3 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.9 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.4 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0032771 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.0 | 0.2 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.4 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.7 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.1 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.0 | 0.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.9 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.5 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.5 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 0.3 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.4 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.0 | 0.6 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 | 0.1 | GO:0070510 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.0 | 0.2 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.2 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.2 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.1 | GO:0042097 | interleukin-4 biosynthetic process(GO:0042097) positive regulation of interleukin-10 biosynthetic process(GO:0045082) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.3 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.6 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.8 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.6 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.0 | 0.2 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.2 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.0 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 2.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.3 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.5 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.2 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.0 | 0.5 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.0 | 0.1 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.0 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.4 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.6 | GO:0031076 | embryonic camera-type eye development(GO:0031076) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 0.8 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.5 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.1 | 0.5 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.9 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 2.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.5 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.1 | 0.5 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 1.0 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.8 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 1.0 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 1.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.3 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.8 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 1.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.4 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 2.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.6 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.3 | 0.9 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.2 | 0.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 0.5 | GO:0070025 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.2 | 0.5 | GO:0070283 | lipoate synthase activity(GO:0016992) radical SAM enzyme activity(GO:0070283) |
| 0.2 | 0.5 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.1 | 0.4 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.1 | 3.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 2.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 1.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.4 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 0.6 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.3 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 1.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.4 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 1.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.9 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.3 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.1 | 0.3 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.3 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 0.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 1.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.5 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.2 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.9 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.4 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 1.1 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.7 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.5 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 1.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.4 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 2.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 1.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.8 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.0 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 1.2 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 0.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |