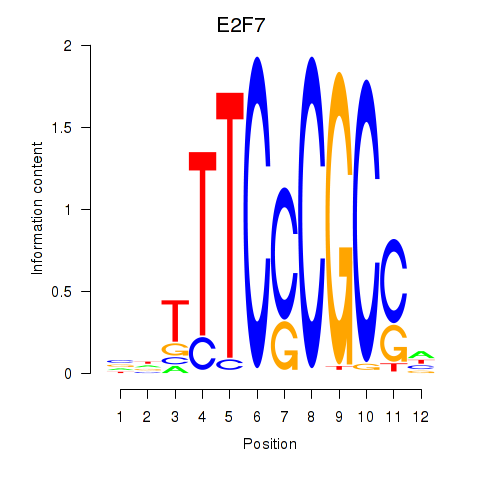
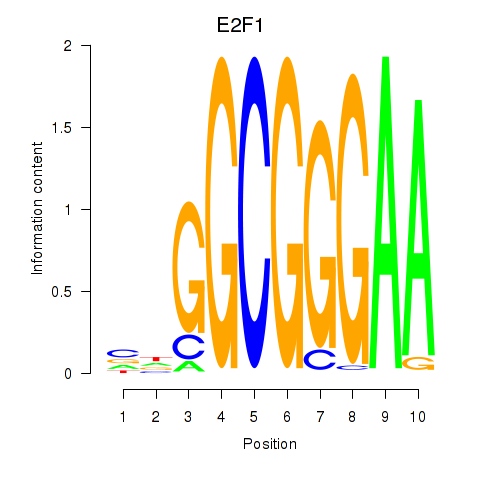
| 0.8 |
2.5 |
GO:0032877 |
positive regulation of DNA endoreduplication(GO:0032877) |
| 0.4 |
1.5 |
GO:0019087 |
transformation of host cell by virus(GO:0019087) |
| 0.3 |
1.0 |
GO:1904772 |
hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.3 |
1.3 |
GO:0046104 |
thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.3 |
3.1 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.3 |
1.1 |
GO:0046452 |
dihydrofolate metabolic process(GO:0046452) |
| 0.3 |
0.8 |
GO:1902299 |
pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.2 |
2.0 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.2 |
0.7 |
GO:0048627 |
myoblast development(GO:0048627) |
| 0.2 |
0.6 |
GO:0000706 |
meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.2 |
1.2 |
GO:0000727 |
double-strand break repair via break-induced replication(GO:0000727) |
| 0.2 |
0.5 |
GO:0018262 |
isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.1 |
0.7 |
GO:0072719 |
cellular response to cisplatin(GO:0072719) |
| 0.1 |
1.5 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 |
0.4 |
GO:0031508 |
pericentric heterochromatin assembly(GO:0031508) |
| 0.1 |
0.3 |
GO:0006272 |
leading strand elongation(GO:0006272) |
| 0.1 |
0.6 |
GO:0030200 |
heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 |
0.5 |
GO:0035978 |
mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) oncogene-induced cell senescence(GO:0090402) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.1 |
0.4 |
GO:0070512 |
regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.1 |
0.7 |
GO:0072180 |
mesonephric duct morphogenesis(GO:0072180) |
| 0.1 |
1.2 |
GO:0051096 |
positive regulation of helicase activity(GO:0051096) |
| 0.1 |
0.4 |
GO:0018312 |
peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.1 |
0.8 |
GO:0098535 |
de novo centriole assembly(GO:0098535) |
| 0.1 |
0.8 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 |
2.6 |
GO:0009263 |
deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 |
0.7 |
GO:0014038 |
regulation of Schwann cell differentiation(GO:0014038) |
| 0.1 |
0.2 |
GO:0014859 |
negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 |
0.6 |
GO:0042816 |
vitamin B6 metabolic process(GO:0042816) |
| 0.1 |
0.4 |
GO:0043504 |
mitochondrial DNA repair(GO:0043504) |
| 0.1 |
4.5 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.1 |
0.3 |
GO:1901069 |
guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 |
1.2 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.1 |
0.3 |
GO:1904956 |
regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.1 |
0.3 |
GO:0002023 |
reduction of food intake in response to dietary excess(GO:0002023) |
| 0.1 |
0.3 |
GO:0060381 |
regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.1 |
3.3 |
GO:0031055 |
chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 |
0.3 |
GO:0060061 |
Spemann organizer formation(GO:0060061) |
| 0.1 |
0.5 |
GO:0035407 |
histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 |
0.3 |
GO:0002514 |
B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.1 |
0.1 |
GO:0002582 |
positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.1 |
0.2 |
GO:0009786 |
regulation of asymmetric cell division(GO:0009786) |
| 0.1 |
0.6 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 |
0.7 |
GO:0060235 |
lens induction in camera-type eye(GO:0060235) |
| 0.1 |
0.1 |
GO:0072054 |
renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.1 |
0.1 |
GO:0003134 |
BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.1 |
0.2 |
GO:0014707 |
branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 |
0.3 |
GO:0045875 |
negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 |
0.2 |
GO:1904747 |
apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.1 |
0.3 |
GO:0097021 |
Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 |
0.9 |
GO:1902659 |
regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.1 |
0.2 |
GO:0046586 |
regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 |
0.4 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.1 |
0.1 |
GO:2000137 |
negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 0.1 |
0.2 |
GO:0072095 |
regulation of branch elongation involved in ureteric bud branching(GO:0072095) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 |
0.2 |
GO:0071139 |
resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.1 |
0.1 |
GO:0007070 |
negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.1 |
0.2 |
GO:0071314 |
cellular response to cocaine(GO:0071314) |
| 0.1 |
0.1 |
GO:0060435 |
bronchiole development(GO:0060435) |
| 0.1 |
0.2 |
GO:0015728 |
mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.1 |
0.2 |
GO:1900220 |
semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 |
0.3 |
GO:0048749 |
compound eye development(GO:0048749) |
| 0.1 |
0.5 |
GO:0048014 |
Tie signaling pathway(GO:0048014) |
| 0.1 |
0.8 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.1 |
0.2 |
GO:0007497 |
posterior midgut development(GO:0007497) endothelin receptor signaling pathway(GO:0086100) |
| 0.1 |
0.5 |
GO:0006189 |
'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 |
0.4 |
GO:0006540 |
glutamate decarboxylation to succinate(GO:0006540) |
| 0.1 |
0.2 |
GO:0035425 |
autocrine signaling(GO:0035425) |
| 0.1 |
0.2 |
GO:1903570 |
regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 |
0.2 |
GO:1901675 |
negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 |
0.2 |
GO:0042986 |
positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 |
0.3 |
GO:0071460 |
cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 |
0.2 |
GO:0071344 |
diphosphate metabolic process(GO:0071344) |
| 0.1 |
0.5 |
GO:0033504 |
floor plate development(GO:0033504) |
| 0.1 |
0.7 |
GO:0042659 |
regulation of cell fate specification(GO:0042659) |
| 0.1 |
0.2 |
GO:0072019 |
carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.1 |
0.1 |
GO:0042138 |
meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 |
0.6 |
GO:0061087 |
positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 |
0.3 |
GO:0042631 |
cellular response to water deprivation(GO:0042631) |
| 0.1 |
0.7 |
GO:0000733 |
DNA strand renaturation(GO:0000733) |
| 0.1 |
0.3 |
GO:0045876 |
positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 |
0.5 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.1 |
0.3 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.1 |
0.2 |
GO:0045404 |
positive regulation of interleukin-10 biosynthetic process(GO:0045082) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 |
0.2 |
GO:0052255 |
induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.0 |
0.3 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 |
0.2 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 |
0.1 |
GO:0014034 |
neural crest cell fate commitment(GO:0014034) |
| 0.0 |
0.2 |
GO:0010982 |
regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 |
0.2 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.0 |
0.2 |
GO:1900827 |
maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 |
0.1 |
GO:0061209 |
cell proliferation involved in mesonephros development(GO:0061209) |
| 0.0 |
0.1 |
GO:1903770 |
regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.0 |
0.2 |
GO:0071874 |
cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 |
0.2 |
GO:0034473 |
U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 |
0.2 |
GO:0035105 |
sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 |
0.1 |
GO:1990108 |
protein linear deubiquitination(GO:1990108) |
| 0.0 |
0.1 |
GO:0032771 |
regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.0 |
0.1 |
GO:0006422 |
aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 |
0.2 |
GO:1900155 |
regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.0 |
0.2 |
GO:0006542 |
glutamine biosynthetic process(GO:0006542) |
| 0.0 |
0.1 |
GO:0006106 |
fumarate metabolic process(GO:0006106) |
| 0.0 |
0.2 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.0 |
0.2 |
GO:0050717 |
positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 |
0.2 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.0 |
0.3 |
GO:0036166 |
phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 |
0.2 |
GO:0072675 |
multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 |
0.1 |
GO:0098838 |
reduced folate transmembrane transport(GO:0098838) |
| 0.0 |
0.1 |
GO:0007089 |
traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 |
0.2 |
GO:0061767 |
negative regulation of lung blood pressure(GO:0061767) |
| 0.0 |
0.2 |
GO:0070734 |
histone H3-K27 methylation(GO:0070734) |
| 0.0 |
0.1 |
GO:1902490 |
regulation of sperm capacitation(GO:1902490) |
| 0.0 |
0.9 |
GO:0000732 |
strand displacement(GO:0000732) |
| 0.0 |
0.3 |
GO:0033314 |
mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 |
0.5 |
GO:0044854 |
plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) |
| 0.0 |
0.4 |
GO:0042670 |
retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 |
0.1 |
GO:0048478 |
replication fork protection(GO:0048478) |
| 0.0 |
0.1 |
GO:0021503 |
neural fold bending(GO:0021503) |
| 0.0 |
0.1 |
GO:0042369 |
vitamin D catabolic process(GO:0042369) |
| 0.0 |
0.1 |
GO:1901097 |
negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 |
0.1 |
GO:0060355 |
positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 |
0.1 |
GO:0089700 |
protein kinase D signaling(GO:0089700) |
| 0.0 |
1.0 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 |
0.2 |
GO:0043418 |
homocysteine catabolic process(GO:0043418) |
| 0.0 |
0.2 |
GO:0048807 |
female genitalia morphogenesis(GO:0048807) |
| 0.0 |
0.2 |
GO:0006021 |
inositol biosynthetic process(GO:0006021) |
| 0.0 |
0.1 |
GO:0060424 |
foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 |
0.1 |
GO:0043686 |
co-translational protein modification(GO:0043686) |
| 0.0 |
0.2 |
GO:0030997 |
regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 |
0.2 |
GO:0048254 |
snoRNA localization(GO:0048254) |
| 0.0 |
0.2 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 |
0.2 |
GO:0044332 |
Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.0 |
0.0 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 |
0.1 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.0 |
0.1 |
GO:0061582 |
intestinal epithelial cell migration(GO:0061582) |
| 0.0 |
0.2 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 |
0.1 |
GO:0006059 |
hexitol metabolic process(GO:0006059) |
| 0.0 |
0.1 |
GO:0002545 |
chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.0 |
0.1 |
GO:0046013 |
T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 |
0.2 |
GO:0060754 |
positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 |
0.1 |
GO:0055099 |
glycoprotein transport(GO:0034436) response to high density lipoprotein particle(GO:0055099) |
| 0.0 |
0.2 |
GO:0006072 |
glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 |
0.5 |
GO:1904816 |
positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.0 |
0.0 |
GO:1904430 |
negative regulation of t-circle formation(GO:1904430) |
| 0.0 |
0.0 |
GO:0014043 |
negative regulation of neuron maturation(GO:0014043) |
| 0.0 |
0.2 |
GO:0061669 |
spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 |
0.1 |
GO:0080154 |
regulation of fertilization(GO:0080154) |
| 0.0 |
0.3 |
GO:0032100 |
positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 |
0.1 |
GO:1902896 |
terminal web assembly(GO:1902896) |
| 0.0 |
0.2 |
GO:1904491 |
protein localization to ciliary transition zone(GO:1904491) |
| 0.0 |
0.1 |
GO:0006288 |
base-excision repair, DNA ligation(GO:0006288) |
| 0.0 |
0.2 |
GO:0007023 |
post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 |
0.1 |
GO:2001038 |
regulation of cellular response to drug(GO:2001038) |
| 0.0 |
0.1 |
GO:0009257 |
10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 |
0.1 |
GO:2001037 |
tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.0 |
0.3 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.0 |
0.1 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 |
0.1 |
GO:0044791 |
modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 |
0.1 |
GO:0006740 |
NADPH regeneration(GO:0006740) |
| 0.0 |
0.1 |
GO:1904046 |
negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.0 |
0.1 |
GO:0045113 |
regulation of integrin biosynthetic process(GO:0045113) |
| 0.0 |
0.3 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 |
0.6 |
GO:1901663 |
ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 |
0.2 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 |
0.0 |
GO:0006172 |
ADP biosynthetic process(GO:0006172) nucleoside diphosphate biosynthetic process(GO:0009133) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 |
0.1 |
GO:0006574 |
valine catabolic process(GO:0006574) |
| 0.0 |
0.2 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 |
0.1 |
GO:1901503 |
ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 |
0.1 |
GO:0015917 |
aminophospholipid transport(GO:0015917) regulation of Cdc42 protein signal transduction(GO:0032489) regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 |
0.2 |
GO:0045950 |
regulation of mitotic recombination(GO:0000019) negative regulation of mitotic recombination(GO:0045950) |
| 0.0 |
0.1 |
GO:0048619 |
embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 |
0.1 |
GO:0071030 |
nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 |
0.1 |
GO:1902499 |
positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 |
0.1 |
GO:0046101 |
hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 |
0.1 |
GO:1902534 |
single-organism membrane invagination(GO:1902534) |
| 0.0 |
0.1 |
GO:0021555 |
midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 |
0.1 |
GO:0097089 |
methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 |
0.2 |
GO:0034720 |
histone H3-K4 demethylation(GO:0034720) |
| 0.0 |
0.0 |
GO:1903224 |
regulation of endodermal cell differentiation(GO:1903224) negative regulation of endodermal cell differentiation(GO:1903225) negative regulation of gastrulation(GO:2000542) |
| 0.0 |
0.0 |
GO:2000812 |
regulation of barbed-end actin filament capping(GO:2000812) |
| 0.0 |
0.0 |
GO:0034163 |
regulation of toll-like receptor 9 signaling pathway(GO:0034163) |
| 0.0 |
0.0 |
GO:0071283 |
cellular response to iron(III) ion(GO:0071283) |
| 0.0 |
0.1 |
GO:0043587 |
tongue morphogenesis(GO:0043587) |
| 0.0 |
0.1 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 |
0.1 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.0 |
0.3 |
GO:0044818 |
mitotic G2/M transition checkpoint(GO:0044818) |
| 0.0 |
0.1 |
GO:0051103 |
DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 |
0.1 |
GO:2000283 |
regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 |
0.1 |
GO:1902902 |
negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 |
0.1 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 |
0.5 |
GO:0006337 |
nucleosome disassembly(GO:0006337) |
| 0.0 |
0.6 |
GO:0031116 |
positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 |
0.1 |
GO:0090267 |
positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 |
0.2 |
GO:1904263 |
positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 |
0.0 |
GO:2000547 |
regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 |
0.2 |
GO:0042908 |
xenobiotic transport(GO:0042908) |
| 0.0 |
0.1 |
GO:1904636 |
response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 |
0.1 |
GO:1902303 |
negative regulation of potassium ion export(GO:1902303) |
| 0.0 |
0.1 |
GO:0032455 |
nerve growth factor processing(GO:0032455) |
| 0.0 |
0.2 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 |
0.4 |
GO:0035458 |
cellular response to interferon-beta(GO:0035458) |
| 0.0 |
0.1 |
GO:0071848 |
circadian temperature homeostasis(GO:0060086) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.0 |
0.1 |
GO:0021718 |
superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 |
0.1 |
GO:1903070 |
negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 |
0.1 |
GO:2001034 |
positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 |
0.2 |
GO:0006271 |
DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 |
0.4 |
GO:0000920 |
cell separation after cytokinesis(GO:0000920) |
| 0.0 |
2.3 |
GO:0051225 |
spindle assembly(GO:0051225) |
| 0.0 |
0.2 |
GO:0051388 |
positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 |
0.1 |
GO:2000172 |
regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 |
0.1 |
GO:0034334 |
adherens junction maintenance(GO:0034334) |
| 0.0 |
0.2 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 |
0.2 |
GO:1903800 |
positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 |
0.5 |
GO:0015012 |
heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 |
0.1 |
GO:0044800 |
fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 |
0.1 |
GO:1904044 |
response to aldosterone(GO:1904044) |
| 0.0 |
0.1 |
GO:0097045 |
phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.0 |
0.1 |
GO:1903416 |
response to glycoside(GO:1903416) |
| 0.0 |
0.2 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.3 |
GO:0051481 |
negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 |
0.2 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 |
0.1 |
GO:0043376 |
regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 |
0.1 |
GO:0035610 |
protein side chain deglutamylation(GO:0035610) |
| 0.0 |
0.0 |
GO:0032077 |
positive regulation of deoxyribonuclease activity(GO:0032077) |
| 0.0 |
0.2 |
GO:0045005 |
DNA-dependent DNA replication maintenance of fidelity(GO:0045005) |
| 0.0 |
0.2 |
GO:0010668 |
ectodermal cell differentiation(GO:0010668) |
| 0.0 |
0.1 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 |
0.0 |
GO:0071529 |
cementum mineralization(GO:0071529) |
| 0.0 |
1.5 |
GO:0071349 |
interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 |
0.1 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.0 |
0.1 |
GO:0061732 |
mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 |
0.2 |
GO:0032049 |
cardiolipin biosynthetic process(GO:0032049) |
| 0.0 |
0.1 |
GO:0036444 |
calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 |
0.2 |
GO:0046498 |
S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 |
0.1 |
GO:0014826 |
vein smooth muscle contraction(GO:0014826) |
| 0.0 |
0.1 |
GO:0006311 |
meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.0 |
0.1 |
GO:0010756 |
positive regulation of plasminogen activation(GO:0010756) |
| 0.0 |
0.1 |
GO:2000418 |
positive regulation of eosinophil migration(GO:2000418) |
| 0.0 |
0.1 |
GO:0039534 |
negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 |
0.1 |
GO:1901355 |
response to rapamycin(GO:1901355) response to sodium phosphate(GO:1904383) |
| 0.0 |
0.1 |
GO:1901569 |
leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 |
0.1 |
GO:0090170 |
MAPK import into nucleus(GO:0000189) trachea formation(GO:0060440) regulation of Golgi inheritance(GO:0090170) |
| 0.0 |
0.1 |
GO:0046485 |
ether lipid metabolic process(GO:0046485) |
| 0.0 |
0.1 |
GO:1990637 |
response to prolactin(GO:1990637) |
| 0.0 |
0.3 |
GO:0070986 |
left/right axis specification(GO:0070986) |
| 0.0 |
0.0 |
GO:0046946 |
hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 |
0.1 |
GO:0043335 |
protein unfolding(GO:0043335) |
| 0.0 |
0.1 |
GO:0007135 |
meiosis II(GO:0007135) |
| 0.0 |
0.0 |
GO:0002143 |
tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 |
0.1 |
GO:0051597 |
response to methylmercury(GO:0051597) |
| 0.0 |
0.0 |
GO:0038162 |
erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 |
0.2 |
GO:0098789 |
pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 |
0.2 |
GO:0090168 |
Golgi reassembly(GO:0090168) |
| 0.0 |
0.1 |
GO:0030421 |
defecation(GO:0030421) |
| 0.0 |
0.0 |
GO:1990785 |
response to water-immersion restraint stress(GO:1990785) |
| 0.0 |
0.2 |
GO:0090385 |
phagosome-lysosome fusion(GO:0090385) |
| 0.0 |
0.1 |
GO:0016075 |
rRNA catabolic process(GO:0016075) |
| 0.0 |
0.1 |
GO:0098967 |
exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 |
0.2 |
GO:0031572 |
G2 DNA damage checkpoint(GO:0031572) |
| 0.0 |
0.1 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 |
0.0 |
GO:0061114 |
hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.0 |
0.6 |
GO:0043928 |
exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 |
0.2 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.1 |
GO:0060849 |
radial pattern formation(GO:0009956) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 |
0.1 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 |
0.2 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.0 |
0.0 |
GO:1904016 |
response to Thyroglobulin triiodothyronine(GO:1904016) |
| 0.0 |
0.0 |
GO:0002184 |
cytoplasmic translational termination(GO:0002184) |
| 0.0 |
0.4 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.2 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 |
0.3 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 |
0.1 |
GO:0030423 |
targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 |
0.3 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 |
0.0 |
GO:0007343 |
egg activation(GO:0007343) |
| 0.0 |
0.1 |
GO:0060392 |
negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 |
0.1 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 |
0.4 |
GO:0051310 |
metaphase plate congression(GO:0051310) |
| 0.0 |
0.1 |
GO:0070317 |
negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 |
0.1 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 |
0.1 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 |
0.1 |
GO:1903974 |
mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.0 |
0.0 |
GO:0009258 |
10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 |
0.1 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.0 |
0.2 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.1 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.0 |
0.3 |
GO:0051016 |
barbed-end actin filament capping(GO:0051016) |
| 0.0 |
0.1 |
GO:0006741 |
NADP biosynthetic process(GO:0006741) |
| 0.0 |
0.2 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.0 |
0.0 |
GO:0000393 |
spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 |
0.2 |
GO:0043584 |
nose development(GO:0043584) |
| 0.0 |
0.0 |
GO:0060352 |
cell adhesion molecule production(GO:0060352) |
| 0.0 |
0.1 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.0 |
0.1 |
GO:1904322 |
response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 |
0.1 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 |
0.3 |
GO:0016254 |
preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 |
0.1 |
GO:2000504 |
positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 |
0.1 |
GO:0061430 |
bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.0 |
0.0 |
GO:1901842 |
negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 |
0.3 |
GO:0036150 |
phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 |
0.2 |
GO:0034199 |
activation of protein kinase A activity(GO:0034199) |
| 0.0 |
0.2 |
GO:0071173 |
mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.0 |
0.0 |
GO:0048631 |
regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 |
0.1 |
GO:0021943 |
formation of radial glial scaffolds(GO:0021943) |