Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for EN2_GBX2_LBX2
Z-value: 0.36
Transcription factors associated with EN2_GBX2_LBX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
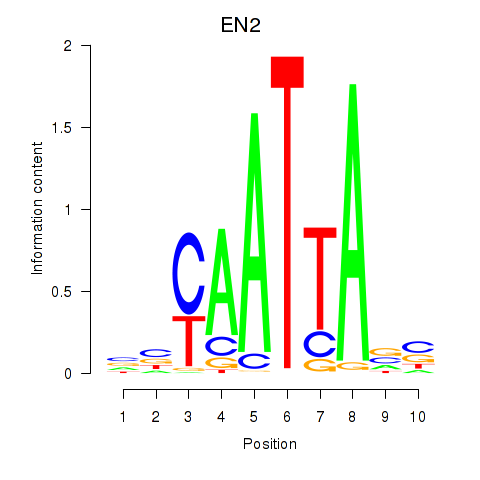
EN2
|
ENSG00000164778.4 | engrailed homeobox 2 |
|
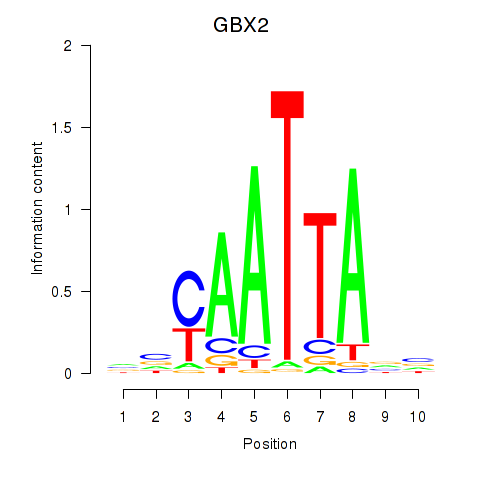
GBX2
|
ENSG00000168505.6 | gastrulation brain homeobox 2 |
|
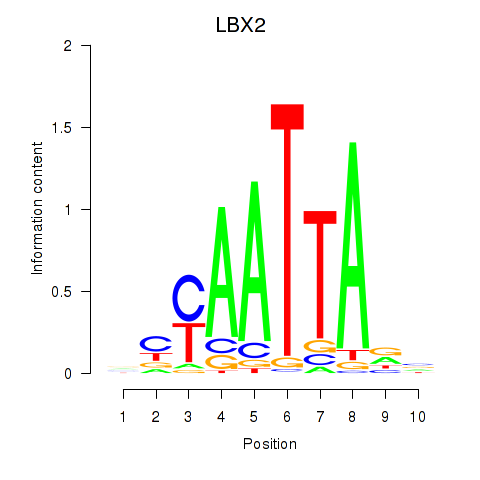
LBX2
|
ENSG00000179528.11 | ladybird homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EN2 | hg19_v2_chr7_+_155250824_155250824 | 0.10 | 6.3e-01 | Click! |
| GBX2 | hg19_v2_chr2_-_237076992_237077012 | 0.05 | 8.1e-01 | Click! |
Activity profile of EN2_GBX2_LBX2 motif
Sorted Z-values of EN2_GBX2_LBX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_71959159 | 1.21 |
ENST00000494131.2
ENST00000397914.4 ENST00000340533.4 |
CYB5A
|
cytochrome b5 type A (microsomal) |
| chr5_-_95297534 | 0.73 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr15_-_55562479 | 0.72 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr5_+_125758865 | 0.68 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr5_+_125758813 | 0.68 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr15_-_55563072 | 0.67 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr15_-_55562582 | 0.63 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr5_-_95297678 | 0.58 |
ENST00000237853.4
|
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr18_+_59000815 | 0.56 |
ENST00000262717.4
|
CDH20
|
cadherin 20, type 2 |
| chr18_-_33709268 | 0.47 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr18_+_55888767 | 0.44 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr20_+_18488137 | 0.38 |
ENST00000450074.1
ENST00000262544.2 ENST00000336714.3 ENST00000377475.3 |
SEC23B
|
Sec23 homolog B (S. cerevisiae) |
| chr1_-_118472171 | 0.36 |
ENST00000369442.3
|
GDAP2
|
ganglioside induced differentiation associated protein 2 |
| chr7_-_92777606 | 0.36 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr3_+_130569429 | 0.33 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr3_+_133292759 | 0.30 |
ENST00000431519.2
|
CDV3
|
CDV3 homolog (mouse) |
| chr7_-_122339162 | 0.30 |
ENST00000340112.2
|
RNF133
|
ring finger protein 133 |
| chr8_-_10512569 | 0.28 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr14_-_51027838 | 0.28 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr18_+_32173276 | 0.27 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr11_-_40315640 | 0.26 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr1_+_101003687 | 0.26 |
ENST00000315033.4
|
GPR88
|
G protein-coupled receptor 88 |
| chr6_+_135502501 | 0.25 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr3_+_111717511 | 0.24 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr3_+_111717600 | 0.24 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr4_-_39979576 | 0.24 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr4_-_105416039 | 0.24 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr1_+_160160283 | 0.23 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr17_-_39203519 | 0.22 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr1_+_160160346 | 0.22 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr17_-_38911580 | 0.22 |
ENST00000312150.4
|
KRT25
|
keratin 25 |
| chr5_+_66300446 | 0.22 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr6_+_26199737 | 0.21 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chr18_-_33702078 | 0.21 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr3_+_111718036 | 0.21 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chrM_+_12331 | 0.21 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr5_-_35938674 | 0.20 |
ENST00000397366.1
ENST00000513623.1 ENST00000514524.1 ENST00000397367.2 |
CAPSL
|
calcyphosine-like |
| chrX_-_21676442 | 0.20 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr6_-_31107127 | 0.19 |
ENST00000259845.4
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chr18_-_31803435 | 0.19 |
ENST00000589544.1
ENST00000269185.4 ENST00000261592.5 |
NOL4
|
nucleolar protein 4 |
| chr11_+_75526212 | 0.19 |
ENST00000356136.3
|
UVRAG
|
UV radiation resistance associated |
| chr3_+_115342349 | 0.19 |
ENST00000393780.3
|
GAP43
|
growth associated protein 43 |
| chr17_+_59489112 | 0.18 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
| chr12_-_56694142 | 0.18 |
ENST00000550655.1
ENST00000548567.1 ENST00000551430.2 ENST00000351328.3 |
CS
|
citrate synthase |
| chr19_+_7030589 | 0.16 |
ENST00000329753.5
|
MBD3L5
|
methyl-CpG binding domain protein 3-like 5 |
| chr3_-_195538728 | 0.15 |
ENST00000349607.4
ENST00000346145.4 |
MUC4
|
mucin 4, cell surface associated |
| chr11_+_57435219 | 0.14 |
ENST00000527985.1
ENST00000287169.3 |
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chr6_-_26199471 | 0.14 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr18_-_31803169 | 0.14 |
ENST00000590712.1
|
NOL4
|
nucleolar protein 4 |
| chr12_-_89746173 | 0.14 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr3_+_111718173 | 0.14 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr8_+_50824233 | 0.13 |
ENST00000522124.1
|
SNTG1
|
syntrophin, gamma 1 |
| chr17_-_39211463 | 0.13 |
ENST00000542910.1
ENST00000398477.1 |
KRTAP2-2
|
keratin associated protein 2-2 |
| chr19_+_51728316 | 0.13 |
ENST00000436584.2
ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33
|
CD33 molecule |
| chr3_+_173116225 | 0.13 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr2_+_65216462 | 0.12 |
ENST00000234256.3
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr1_+_171227069 | 0.12 |
ENST00000354841.4
|
FMO1
|
flavin containing monooxygenase 1 |
| chr19_-_6737576 | 0.12 |
ENST00000601716.1
ENST00000264080.7 |
GPR108
|
G protein-coupled receptor 108 |
| chrX_-_46187069 | 0.11 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr20_-_18447667 | 0.11 |
ENST00000262547.5
ENST00000329494.5 ENST00000357236.4 |
DZANK1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr13_-_36788718 | 0.11 |
ENST00000317764.6
ENST00000379881.3 |
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr3_-_157221380 | 0.11 |
ENST00000468233.1
|
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr14_+_32798462 | 0.11 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr4_+_169013666 | 0.10 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr14_-_82000140 | 0.10 |
ENST00000555824.1
ENST00000557372.1 ENST00000336735.4 |
SEL1L
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr19_-_6737241 | 0.10 |
ENST00000430424.4
ENST00000597298.1 |
GPR108
|
G protein-coupled receptor 108 |
| chr22_+_39916558 | 0.10 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr10_-_50970322 | 0.10 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr15_+_89631381 | 0.10 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chr15_+_89631647 | 0.10 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr10_-_50970382 | 0.10 |
ENST00000419399.1
ENST00000432695.1 |
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr2_-_61697862 | 0.10 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr21_-_32253874 | 0.09 |
ENST00000332378.4
|
KRTAP11-1
|
keratin associated protein 11-1 |
| chr19_+_48949030 | 0.09 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr7_-_73038822 | 0.09 |
ENST00000414749.2
ENST00000429400.2 ENST00000434326.1 |
MLXIPL
|
MLX interacting protein-like |
| chr7_-_73038867 | 0.09 |
ENST00000313375.3
ENST00000354613.1 ENST00000395189.1 ENST00000453275.1 |
MLXIPL
|
MLX interacting protein-like |
| chr4_-_89205705 | 0.09 |
ENST00000295908.7
ENST00000510548.2 ENST00000508256.1 |
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr1_-_152386732 | 0.09 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chr8_+_22424551 | 0.09 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr3_+_172468472 | 0.09 |
ENST00000232458.5
ENST00000392692.3 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr17_-_39306054 | 0.09 |
ENST00000343246.4
|
KRTAP4-5
|
keratin associated protein 4-5 |
| chr8_+_105235572 | 0.09 |
ENST00000523362.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr3_+_172468505 | 0.08 |
ENST00000427830.1
ENST00000417960.1 ENST00000428567.1 ENST00000366090.2 ENST00000426894.1 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr12_+_53662073 | 0.08 |
ENST00000553219.1
ENST00000257934.4 |
ESPL1
|
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr12_+_53662110 | 0.08 |
ENST00000552462.1
|
ESPL1
|
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr6_-_26199499 | 0.08 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr13_-_41593425 | 0.08 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr12_-_16759711 | 0.08 |
ENST00000447609.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr2_+_171034646 | 0.08 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr1_-_23670752 | 0.08 |
ENST00000302271.6
ENST00000426846.2 ENST00000427764.2 ENST00000606561.1 ENST00000374616.3 |
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr2_-_224467093 | 0.08 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr6_-_167040731 | 0.07 |
ENST00000265678.4
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr3_+_152879985 | 0.07 |
ENST00000323534.2
|
RAP2B
|
RAP2B, member of RAS oncogene family |
| chr3_-_119813264 | 0.07 |
ENST00000264235.8
|
GSK3B
|
glycogen synthase kinase 3 beta |
| chr1_-_118472216 | 0.07 |
ENST00000369443.5
|
GDAP2
|
ganglioside induced differentiation associated protein 2 |
| chr3_-_195538760 | 0.07 |
ENST00000475231.1
|
MUC4
|
mucin 4, cell surface associated |
| chr20_+_18447771 | 0.07 |
ENST00000377603.4
|
POLR3F
|
polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa |
| chr19_-_7040190 | 0.07 |
ENST00000381394.4
|
MBD3L4
|
methyl-CpG binding domain protein 3-like 4 |
| chr1_-_23670817 | 0.06 |
ENST00000478691.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr8_-_25281747 | 0.06 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr8_+_92261516 | 0.06 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr19_-_41870026 | 0.06 |
ENST00000243578.3
|
B9D2
|
B9 protein domain 2 |
| chr1_-_23670813 | 0.06 |
ENST00000374612.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr19_-_7698599 | 0.06 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr4_-_89205879 | 0.06 |
ENST00000608933.1
ENST00000315194.4 ENST00000514204.1 |
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr7_-_137028498 | 0.06 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr19_+_45394477 | 0.06 |
ENST00000252487.5
ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr4_-_120243545 | 0.06 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr3_-_52090461 | 0.06 |
ENST00000296483.6
ENST00000495880.1 |
DUSP7
|
dual specificity phosphatase 7 |
| chr9_+_1050331 | 0.06 |
ENST00000382255.3
ENST00000382251.3 ENST00000412350.2 |
DMRT2
|
doublesex and mab-3 related transcription factor 2 |
| chr12_+_7014126 | 0.06 |
ENST00000415834.1
ENST00000436789.1 |
LRRC23
|
leucine rich repeat containing 23 |
| chr1_+_203463245 | 0.05 |
ENST00000367222.2
|
OPTC
|
opticin |
| chr15_-_31393910 | 0.05 |
ENST00000397795.2
ENST00000256552.6 ENST00000559179.1 |
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr9_+_80912059 | 0.05 |
ENST00000347159.2
ENST00000376588.3 |
PSAT1
|
phosphoserine aminotransferase 1 |
| chr6_-_100912785 | 0.05 |
ENST00000369208.3
|
SIM1
|
single-minded family bHLH transcription factor 1 |
| chr9_-_136933615 | 0.05 |
ENST00000371834.2
|
BRD3
|
bromodomain containing 3 |
| chr6_+_131571535 | 0.05 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr20_-_50722183 | 0.05 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr12_-_28123206 | 0.05 |
ENST00000542963.1
ENST00000535992.1 |
PTHLH
|
parathyroid hormone-like hormone |
| chr4_-_174255400 | 0.05 |
ENST00000506267.1
|
HMGB2
|
high mobility group box 2 |
| chr1_-_17766198 | 0.05 |
ENST00000375436.4
|
RCC2
|
regulator of chromosome condensation 2 |
| chr11_-_71823266 | 0.05 |
ENST00000538919.1
ENST00000539395.1 ENST00000542531.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr6_+_34204642 | 0.05 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chrX_+_95939638 | 0.05 |
ENST00000373061.3
ENST00000373054.4 ENST00000355827.4 |
DIAPH2
|
diaphanous-related formin 2 |
| chr1_-_2461684 | 0.05 |
ENST00000378453.3
|
HES5
|
hes family bHLH transcription factor 5 |
| chr16_+_69345243 | 0.05 |
ENST00000254950.11
|
VPS4A
|
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chrX_+_135730297 | 0.05 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr17_-_47045949 | 0.04 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide |
| chr5_-_10761206 | 0.04 |
ENST00000432074.2
ENST00000230895.6 |
DAP
|
death-associated protein |
| chr1_-_152131703 | 0.04 |
ENST00000316073.3
|
RPTN
|
repetin |
| chr1_+_62901968 | 0.04 |
ENST00000452143.1
ENST00000442679.1 ENST00000371146.1 |
USP1
|
ubiquitin specific peptidase 1 |
| chr11_-_71823796 | 0.04 |
ENST00000545680.1
ENST00000543587.1 ENST00000538393.1 ENST00000535234.1 ENST00000227618.4 ENST00000535503.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr1_+_42846443 | 0.04 |
ENST00000410070.2
ENST00000431473.3 |
RIMKLA
|
ribosomal modification protein rimK-like family member A |
| chr2_+_158114051 | 0.04 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr4_+_89206076 | 0.04 |
ENST00000500009.2
|
RP11-10L7.1
|
RP11-10L7.1 |
| chr9_+_12693336 | 0.04 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr4_-_116034979 | 0.04 |
ENST00000264363.2
|
NDST4
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
| chr4_+_144354644 | 0.04 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr11_-_71823715 | 0.04 |
ENST00000545944.1
ENST00000502597.2 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr2_-_163008903 | 0.04 |
ENST00000418842.2
ENST00000375497.3 |
GCG
|
glucagon |
| chr17_-_10017864 | 0.03 |
ENST00000323816.4
|
GAS7
|
growth arrest-specific 7 |
| chr1_+_179923873 | 0.03 |
ENST00000367607.3
ENST00000491495.2 |
CEP350
|
centrosomal protein 350kDa |
| chr12_+_7014064 | 0.03 |
ENST00000443597.2
|
LRRC23
|
leucine rich repeat containing 23 |
| chr19_-_51522955 | 0.03 |
ENST00000358789.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr15_-_64673630 | 0.03 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr1_+_118472343 | 0.03 |
ENST00000369441.3
ENST00000349139.5 |
WDR3
|
WD repeat domain 3 |
| chr3_+_35722487 | 0.03 |
ENST00000441454.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr10_-_56560939 | 0.03 |
ENST00000373955.1
|
PCDH15
|
protocadherin-related 15 |
| chr15_-_64673665 | 0.03 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr8_+_24298597 | 0.03 |
ENST00000380789.1
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr6_-_26032288 | 0.03 |
ENST00000244661.2
|
HIST1H3B
|
histone cluster 1, H3b |
| chr6_-_31782813 | 0.03 |
ENST00000375654.4
|
HSPA1L
|
heat shock 70kDa protein 1-like |
| chr17_-_9929581 | 0.03 |
ENST00000437099.2
ENST00000396115.2 |
GAS7
|
growth arrest-specific 7 |
| chr8_-_18744528 | 0.03 |
ENST00000523619.1
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr1_+_40713573 | 0.03 |
ENST00000372766.3
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr19_-_7968427 | 0.03 |
ENST00000539278.1
|
AC010336.1
|
Uncharacterized protein |
| chr17_+_47448102 | 0.03 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr12_+_7013897 | 0.03 |
ENST00000007969.8
ENST00000323702.5 |
LRRC23
|
leucine rich repeat containing 23 |
| chr2_+_196313239 | 0.03 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr12_-_52828147 | 0.02 |
ENST00000252245.5
|
KRT75
|
keratin 75 |
| chr12_-_101604185 | 0.02 |
ENST00000536262.2
|
SLC5A8
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 8 |
| chr8_+_24298531 | 0.02 |
ENST00000175238.6
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr19_-_19302931 | 0.02 |
ENST00000444486.3
ENST00000514819.3 ENST00000585679.1 ENST00000162023.5 |
MEF2BNB-MEF2B
MEF2BNB
MEF2B
|
MEF2BNB-MEF2B readthrough MEF2B neighbor myocyte enhancer factor 2B |
| chr3_-_141747950 | 0.02 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr17_-_64225508 | 0.02 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr2_-_77749474 | 0.02 |
ENST00000409093.1
ENST00000409088.3 |
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr5_+_31193847 | 0.02 |
ENST00000514738.1
ENST00000265071.2 |
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr12_-_28122980 | 0.02 |
ENST00000395868.3
ENST00000534890.1 |
PTHLH
|
parathyroid hormone-like hormone |
| chr4_+_71458012 | 0.02 |
ENST00000449493.2
|
AMBN
|
ameloblastin (enamel matrix protein) |
| chrX_+_100353153 | 0.02 |
ENST00000423383.1
ENST00000218507.5 ENST00000403304.2 ENST00000435570.1 |
CENPI
|
centromere protein I |
| chr5_+_145826867 | 0.02 |
ENST00000296702.5
ENST00000394421.2 |
TCERG1
|
transcription elongation regulator 1 |
| chr11_-_13517565 | 0.02 |
ENST00000282091.1
ENST00000529816.1 |
PTH
|
parathyroid hormone |
| chr15_+_40733387 | 0.02 |
ENST00000416165.1
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr17_-_48785216 | 0.02 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr6_-_161695042 | 0.02 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr15_+_34638066 | 0.02 |
ENST00000333756.4
|
NUTM1
|
NUT midline carcinoma, family member 1 |
| chr1_+_160370344 | 0.02 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr1_+_110881945 | 0.02 |
ENST00000602849.1
ENST00000487146.2 |
RBM15
|
RNA binding motif protein 15 |
| chr7_-_99717463 | 0.01 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr7_-_25268104 | 0.01 |
ENST00000222674.2
|
NPVF
|
neuropeptide VF precursor |
| chr6_-_139613269 | 0.01 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chrX_+_154444643 | 0.01 |
ENST00000286428.5
|
VBP1
|
von Hippel-Lindau binding protein 1 |
| chr6_-_72130472 | 0.01 |
ENST00000426635.2
|
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr7_-_137028534 | 0.01 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr8_-_42234745 | 0.01 |
ENST00000220812.2
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr6_-_161695074 | 0.01 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr1_+_152943122 | 0.01 |
ENST00000328051.2
|
SPRR4
|
small proline-rich protein 4 |
| chrM_+_7586 | 0.01 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr7_+_129984630 | 0.01 |
ENST00000355388.3
ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5
|
carboxypeptidase A5 |
| chr4_-_174255536 | 0.01 |
ENST00000446922.2
|
HMGB2
|
high mobility group box 2 |
| chr15_+_76629064 | 0.01 |
ENST00000290759.4
|
ISL2
|
ISL LIM homeobox 2 |
| chr4_-_74486217 | 0.01 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr6_+_26020672 | 0.00 |
ENST00000357647.3
|
HIST1H3A
|
histone cluster 1, H3a |
| chr10_+_99205894 | 0.00 |
ENST00000370854.3
ENST00000393760.1 ENST00000414567.1 ENST00000370846.4 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chrX_+_95939711 | 0.00 |
ENST00000373049.4
ENST00000324765.8 |
DIAPH2
|
diaphanous-related formin 2 |
| chr11_-_124190184 | 0.00 |
ENST00000357438.2
|
OR8D2
|
olfactory receptor, family 8, subfamily D, member 2 |
| chr5_+_179159813 | 0.00 |
ENST00000292599.3
|
MAML1
|
mastermind-like 1 (Drosophila) |
| chr5_+_176811431 | 0.00 |
ENST00000512593.1
ENST00000324417.5 |
SLC34A1
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 1 |
| chr10_+_99205959 | 0.00 |
ENST00000352634.4
ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr3_-_157217328 | 0.00 |
ENST00000392832.2
ENST00000543418.1 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of EN2_GBX2_LBX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.3 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.1 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 1.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 0.3 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.4 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.0 | 0.1 | GO:1904339 | superior temporal gyrus development(GO:0071109) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.0 | GO:2000974 | auditory receptor cell fate determination(GO:0042668) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.2 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.2 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.0 | 1.3 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.4 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 1.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 1.2 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0015184 | L-alanine transmembrane transporter activity(GO:0015180) L-cystine transmembrane transporter activity(GO:0015184) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.0 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |