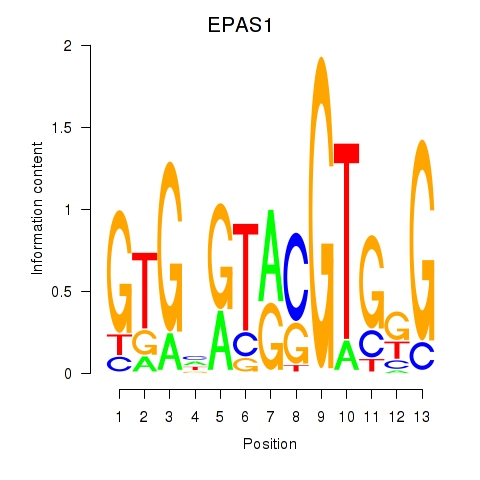
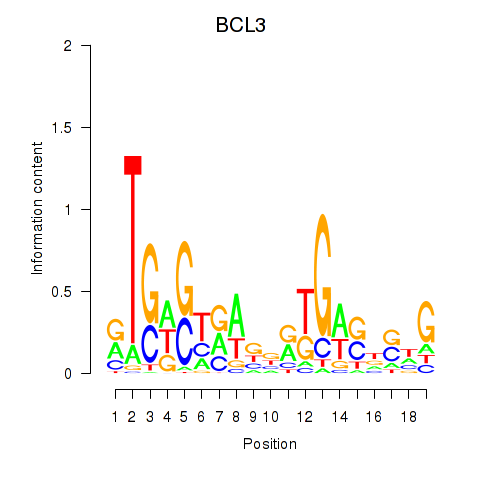
Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for EPAS1_BCL3
Z-value: 1.24
Transcription factors associated with EPAS1_BCL3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EPAS1
|
ENSG00000116016.9 | endothelial PAS domain protein 1 |
|
BCL3
|
ENSG00000069399.8 | BCL3 transcription coactivator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EPAS1 | hg19_v2_chr2_+_46524537_46524553 | 0.52 | 7.4e-03 | Click! |
| BCL3 | hg19_v2_chr19_+_45254529_45254578 | 0.45 | 2.5e-02 | Click! |
Activity profile of EPAS1_BCL3 motif
Sorted Z-values of EPAS1_BCL3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_43964055 | 3.14 |
ENST00000528572.1
|
C11orf96
|
chromosome 11 open reading frame 96 |
| chr20_-_36793663 | 2.74 |
ENST00000536701.1
ENST00000536724.1 |
TGM2
|
transglutaminase 2 |
| chr3_-_18466026 | 2.54 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr7_-_132261253 | 2.07 |
ENST00000321063.4
|
PLXNA4
|
plexin A4 |
| chr15_-_69113218 | 1.96 |
ENST00000560303.1
ENST00000465139.2 |
ANP32A
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr4_-_186732048 | 1.95 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr8_-_81083890 | 1.94 |
ENST00000518937.1
|
TPD52
|
tumor protein D52 |
| chr1_+_110453109 | 1.92 |
ENST00000525659.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr1_+_61547894 | 1.90 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr2_-_188312971 | 1.82 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr20_+_34680620 | 1.76 |
ENST00000430276.1
ENST00000373950.2 ENST00000452261.1 |
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr14_-_92413353 | 1.73 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr3_-_52479043 | 1.71 |
ENST00000231721.2
ENST00000475739.1 |
SEMA3G
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G |
| chr1_+_110453608 | 1.66 |
ENST00000369801.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr3_-_32022733 | 1.61 |
ENST00000438237.2
ENST00000396556.2 |
OSBPL10
|
oxysterol binding protein-like 10 |
| chr4_+_41361616 | 1.54 |
ENST00000513024.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_+_61548225 | 1.52 |
ENST00000371187.3
|
NFIA
|
nuclear factor I/A |
| chr4_-_70626314 | 1.49 |
ENST00000510821.1
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr4_+_140586922 | 1.47 |
ENST00000265498.1
ENST00000506797.1 |
MGST2
|
microsomal glutathione S-transferase 2 |
| chr1_+_110453462 | 1.46 |
ENST00000488198.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr14_-_54420133 | 1.44 |
ENST00000559501.1
ENST00000558984.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr6_-_40555176 | 1.43 |
ENST00000338305.6
|
LRFN2
|
leucine rich repeat and fibronectin type III domain containing 2 |
| chr2_-_238499303 | 1.42 |
ENST00000409576.1
|
RAB17
|
RAB17, member RAS oncogene family |
| chr22_-_37880543 | 1.35 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr2_+_102686820 | 1.31 |
ENST00000409929.1
ENST00000424272.1 |
IL1R1
|
interleukin 1 receptor, type I |
| chr3_-_168865522 | 1.31 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr7_-_150329421 | 1.29 |
ENST00000493969.1
ENST00000328902.5 |
GIMAP6
|
GTPase, IMAP family member 6 |
| chr3_-_142607740 | 1.29 |
ENST00000485766.1
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr20_-_36793774 | 1.28 |
ENST00000361475.2
|
TGM2
|
transglutaminase 2 |
| chr7_+_150688083 | 1.21 |
ENST00000297494.3
|
NOS3
|
nitric oxide synthase 3 (endothelial cell) |
| chr19_-_42806723 | 1.21 |
ENST00000262890.3
|
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr19_+_30863271 | 1.21 |
ENST00000355537.3
|
ZNF536
|
zinc finger protein 536 |
| chr8_-_89339705 | 1.19 |
ENST00000286614.6
|
MMP16
|
matrix metallopeptidase 16 (membrane-inserted) |
| chr7_+_150688166 | 1.18 |
ENST00000461406.1
|
NOS3
|
nitric oxide synthase 3 (endothelial cell) |
| chr9_+_34652164 | 1.16 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr8_+_21915368 | 1.15 |
ENST00000265800.5
ENST00000517418.1 |
DMTN
|
dematin actin binding protein |
| chr4_-_70626430 | 1.14 |
ENST00000310613.3
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr4_-_102268484 | 1.14 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr13_+_114238997 | 1.13 |
ENST00000538138.1
ENST00000375370.5 |
TFDP1
|
transcription factor Dp-1 |
| chr2_+_20646824 | 1.12 |
ENST00000272233.4
|
RHOB
|
ras homolog family member B |
| chr4_-_102268628 | 1.12 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr1_+_61548374 | 1.07 |
ENST00000485903.2
ENST00000371185.2 ENST00000371184.2 |
NFIA
|
nuclear factor I/A |
| chr6_-_36807762 | 1.02 |
ENST00000244751.2
|
CPNE5
|
copine V |
| chr17_-_7080227 | 1.02 |
ENST00000574330.1
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr6_-_16761678 | 0.98 |
ENST00000244769.4
ENST00000436367.1 |
ATXN1
|
ataxin 1 |
| chr4_+_41362796 | 0.97 |
ENST00000508501.1
ENST00000512946.1 ENST00000313860.7 ENST00000512632.1 ENST00000512820.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr7_+_136553370 | 0.96 |
ENST00000445907.2
|
CHRM2
|
cholinergic receptor, muscarinic 2 |
| chr14_-_71107921 | 0.96 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr4_+_156588806 | 0.95 |
ENST00000513574.1
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr21_+_46117087 | 0.92 |
ENST00000400365.3
|
KRTAP10-12
|
keratin associated protein 10-12 |
| chr19_+_45973120 | 0.91 |
ENST00000592811.1
ENST00000586615.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr18_+_77155856 | 0.91 |
ENST00000253506.5
ENST00000591814.1 |
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr8_-_81083341 | 0.91 |
ENST00000519303.2
|
TPD52
|
tumor protein D52 |
| chr14_+_105047478 | 0.91 |
ENST00000410013.1
|
C14orf180
|
chromosome 14 open reading frame 180 |
| chr19_+_1407733 | 0.91 |
ENST00000592453.1
|
DAZAP1
|
DAZ associated protein 1 |
| chr6_+_130686856 | 0.89 |
ENST00000296978.3
|
TMEM200A
|
transmembrane protein 200A |
| chr1_+_203595903 | 0.89 |
ENST00000367218.3
ENST00000367219.3 ENST00000391954.2 |
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr12_+_20522179 | 0.88 |
ENST00000359062.3
|
PDE3A
|
phosphodiesterase 3A, cGMP-inhibited |
| chr11_-_414948 | 0.88 |
ENST00000530494.1
ENST00000528209.1 ENST00000431843.2 ENST00000528058.1 |
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr8_+_68864330 | 0.87 |
ENST00000288368.4
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr1_+_92495528 | 0.86 |
ENST00000370383.4
|
EPHX4
|
epoxide hydrolase 4 |
| chr7_-_50518022 | 0.85 |
ENST00000356889.4
ENST00000420829.1 ENST00000448788.1 ENST00000395556.2 ENST00000422854.1 ENST00000435566.1 ENST00000433017.1 |
FIGNL1
|
fidgetin-like 1 |
| chr3_-_171177852 | 0.84 |
ENST00000284483.8
ENST00000475336.1 ENST00000357327.5 ENST00000460047.1 ENST00000488470.1 ENST00000470834.1 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr3_+_134514093 | 0.84 |
ENST00000398015.3
|
EPHB1
|
EPH receptor B1 |
| chr1_+_203595689 | 0.83 |
ENST00000357681.5
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr17_+_2699697 | 0.83 |
ENST00000254695.8
ENST00000366401.4 ENST00000542807.1 |
RAP1GAP2
|
RAP1 GTPase activating protein 2 |
| chr22_-_39637135 | 0.82 |
ENST00000440375.1
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr5_-_61031495 | 0.81 |
ENST00000506550.1
ENST00000512882.2 |
CTD-2170G1.2
|
CTD-2170G1.2 |
| chr17_+_61571746 | 0.80 |
ENST00000579409.1
|
ACE
|
angiotensin I converting enzyme |
| chr3_-_18466787 | 0.79 |
ENST00000338745.6
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr11_-_89224488 | 0.78 |
ENST00000534731.1
ENST00000527626.1 |
NOX4
|
NADPH oxidase 4 |
| chr11_-_89224508 | 0.76 |
ENST00000525196.1
|
NOX4
|
NADPH oxidase 4 |
| chr1_+_110453203 | 0.76 |
ENST00000357302.4
ENST00000344188.5 ENST00000329608.6 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr8_+_21916710 | 0.76 |
ENST00000523266.1
ENST00000519907.1 |
DMTN
|
dematin actin binding protein |
| chr6_+_38690622 | 0.76 |
ENST00000327475.6
|
DNAH8
|
dynein, axonemal, heavy chain 8 |
| chr17_+_39382900 | 0.76 |
ENST00000377721.3
ENST00000455970.2 |
KRTAP9-2
|
keratin associated protein 9-2 |
| chr12_+_26348582 | 0.75 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr8_+_21916680 | 0.74 |
ENST00000358242.3
ENST00000415253.1 |
DMTN
|
dematin actin binding protein |
| chr17_+_4618734 | 0.72 |
ENST00000571206.1
|
ARRB2
|
arrestin, beta 2 |
| chr2_+_85360499 | 0.72 |
ENST00000282111.3
|
TCF7L1
|
transcription factor 7-like 1 (T-cell specific, HMG-box) |
| chr12_-_49259643 | 0.72 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr4_+_86748898 | 0.72 |
ENST00000509300.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr9_-_39288092 | 0.71 |
ENST00000323947.7
ENST00000297668.6 ENST00000377656.2 ENST00000377659.1 |
CNTNAP3
|
contactin associated protein-like 3 |
| chr9_-_14314518 | 0.68 |
ENST00000397581.2
|
NFIB
|
nuclear factor I/B |
| chr22_-_29075853 | 0.68 |
ENST00000397906.2
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr11_-_89224299 | 0.68 |
ENST00000343727.5
ENST00000531342.1 ENST00000375979.3 |
NOX4
|
NADPH oxidase 4 |
| chr15_+_66994561 | 0.68 |
ENST00000288840.5
|
SMAD6
|
SMAD family member 6 |
| chr2_+_11273179 | 0.67 |
ENST00000381585.3
ENST00000405022.3 |
C2orf50
|
chromosome 2 open reading frame 50 |
| chr6_+_12012536 | 0.67 |
ENST00000379388.2
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr7_+_136553824 | 0.66 |
ENST00000320658.5
ENST00000453373.1 ENST00000397608.3 ENST00000402486.3 ENST00000401861.1 |
CHRM2
|
cholinergic receptor, muscarinic 2 |
| chr11_+_10472223 | 0.65 |
ENST00000396554.3
ENST00000524866.1 |
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr20_+_10199468 | 0.65 |
ENST00000254976.2
ENST00000304886.2 |
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr2_-_220436248 | 0.64 |
ENST00000265318.4
|
OBSL1
|
obscurin-like 1 |
| chr3_-_15540055 | 0.64 |
ENST00000605797.1
ENST00000435459.2 |
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr18_+_77155942 | 0.64 |
ENST00000397790.2
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr21_+_37507210 | 0.64 |
ENST00000290354.5
|
CBR3
|
carbonyl reductase 3 |
| chr22_+_19705928 | 0.63 |
ENST00000383045.3
ENST00000438754.2 |
SEPT5
|
septin 5 |
| chr9_-_72287191 | 0.63 |
ENST00000265381.4
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1 |
| chr3_-_142608001 | 0.63 |
ENST00000295992.3
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr11_-_74022658 | 0.62 |
ENST00000427714.2
ENST00000331597.4 |
P4HA3
|
prolyl 4-hydroxylase, alpha polypeptide III |
| chr11_-_89224638 | 0.61 |
ENST00000535633.1
ENST00000263317.4 |
NOX4
|
NADPH oxidase 4 |
| chr4_-_89152474 | 0.61 |
ENST00000515655.1
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr8_-_145691031 | 0.61 |
ENST00000424149.2
ENST00000530637.1 ENST00000306145.5 |
CYHR1
|
cysteine/histidine-rich 1 |
| chr5_-_39425068 | 0.60 |
ENST00000515700.1
ENST00000339788.6 |
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr12_+_79258547 | 0.60 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr21_-_31312230 | 0.60 |
ENST00000399907.1
ENST00000399909.1 |
GRIK1
|
glutamate receptor, ionotropic, kainate 1 |
| chr1_-_85930246 | 0.60 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr1_+_153651078 | 0.59 |
ENST00000368680.3
|
NPR1
|
natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A) |
| chr19_-_42806842 | 0.59 |
ENST00000596265.1
|
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr6_+_74405501 | 0.58 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr10_-_30348439 | 0.57 |
ENST00000375377.1
|
KIAA1462
|
KIAA1462 |
| chr12_+_79258444 | 0.57 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr22_-_39636914 | 0.57 |
ENST00000381551.4
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr6_-_42690312 | 0.57 |
ENST00000230381.5
|
PRPH2
|
peripherin 2 (retinal degeneration, slow) |
| chr14_+_96505659 | 0.56 |
ENST00000555004.1
|
C14orf132
|
chromosome 14 open reading frame 132 |
| chr9_-_20622478 | 0.55 |
ENST00000355930.6
ENST00000380338.4 |
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr4_-_159080806 | 0.54 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr14_+_61995722 | 0.54 |
ENST00000556347.1
|
RP11-47I22.4
|
RP11-47I22.4 |
| chr2_-_85645545 | 0.54 |
ENST00000409275.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr3_+_37903432 | 0.54 |
ENST00000443503.2
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr11_-_73471655 | 0.53 |
ENST00000400470.2
|
RAB6A
|
RAB6A, member RAS oncogene family |
| chr19_-_42806919 | 0.53 |
ENST00000595530.1
ENST00000538771.1 ENST00000601865.1 |
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr4_+_144258021 | 0.53 |
ENST00000262994.4
|
GAB1
|
GRB2-associated binding protein 1 |
| chr7_-_38398721 | 0.52 |
ENST00000390346.2
|
TRGV3
|
T cell receptor gamma variable 3 |
| chr19_+_36705504 | 0.52 |
ENST00000456324.1
|
ZNF146
|
zinc finger protein 146 |
| chr8_+_10530155 | 0.52 |
ENST00000521818.1
|
C8orf74
|
chromosome 8 open reading frame 74 |
| chr4_-_141075330 | 0.52 |
ENST00000509479.2
|
MAML3
|
mastermind-like 3 (Drosophila) |
| chr6_+_31620191 | 0.51 |
ENST00000375918.2
ENST00000375920.4 |
APOM
|
apolipoprotein M |
| chr4_+_86749045 | 0.51 |
ENST00000514229.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr1_-_44497024 | 0.50 |
ENST00000372306.3
ENST00000372310.3 ENST00000475075.2 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr1_-_44497118 | 0.50 |
ENST00000537678.1
ENST00000466926.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr6_+_74405804 | 0.50 |
ENST00000287097.5
|
CD109
|
CD109 molecule |
| chr2_+_85646054 | 0.50 |
ENST00000389938.2
|
SH2D6
|
SH2 domain containing 6 |
| chr3_+_52009066 | 0.49 |
ENST00000458031.2
ENST00000463937.1 ENST00000273596.3 |
ACY1
ABHD14A-ACY1
ABHD14A
|
aminoacylase 1 ABHD14A-ACY1 readthrough (NMD candidate) abhydrolase domain containing 14A |
| chr3_-_9291063 | 0.49 |
ENST00000383836.3
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr19_+_18530146 | 0.49 |
ENST00000348495.6
ENST00000270061.7 |
SSBP4
|
single stranded DNA binding protein 4 |
| chr1_+_82266053 | 0.49 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr19_+_13106383 | 0.49 |
ENST00000397661.2
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr1_+_86046433 | 0.48 |
ENST00000451137.2
|
CYR61
|
cysteine-rich, angiogenic inducer, 61 |
| chr3_+_186435137 | 0.48 |
ENST00000447445.1
|
KNG1
|
kininogen 1 |
| chr7_+_114562172 | 0.48 |
ENST00000393486.1
ENST00000257724.3 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr18_-_53089723 | 0.48 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chr6_+_36646435 | 0.48 |
ENST00000244741.5
ENST00000405375.1 ENST00000373711.2 |
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
| chr21_-_31311818 | 0.48 |
ENST00000535441.1
ENST00000309434.7 ENST00000327783.4 ENST00000389124.2 ENST00000389125.3 ENST00000399913.1 |
GRIK1
|
glutamate receptor, ionotropic, kainate 1 |
| chr14_-_105420241 | 0.48 |
ENST00000557457.1
|
AHNAK2
|
AHNAK nucleoprotein 2 |
| chr17_-_56406117 | 0.47 |
ENST00000268893.6
ENST00000355701.3 |
BZRAP1
|
benzodiazepine receptor (peripheral) associated protein 1 |
| chr20_-_2781222 | 0.47 |
ENST00000380605.2
|
CPXM1
|
carboxypeptidase X (M14 family), member 1 |
| chr18_+_6729698 | 0.47 |
ENST00000383472.4
|
ARHGAP28
|
Rho GTPase activating protein 28 |
| chrX_+_135229600 | 0.46 |
ENST00000370690.3
|
FHL1
|
four and a half LIM domains 1 |
| chr18_+_61254534 | 0.46 |
ENST00000269489.5
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr7_+_29519486 | 0.46 |
ENST00000409041.4
|
CHN2
|
chimerin 2 |
| chr2_+_219247021 | 0.45 |
ENST00000539932.1
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr6_-_84419101 | 0.45 |
ENST00000520302.1
ENST00000520213.1 ENST00000439399.2 ENST00000428679.2 ENST00000437520.1 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr9_+_43684902 | 0.45 |
ENST00000377564.3
ENST00000276974.6 |
CNTNAP3B
|
contactin associated protein-like 3B |
| chr1_+_77747656 | 0.45 |
ENST00000354567.2
|
AK5
|
adenylate kinase 5 |
| chr9_-_13165457 | 0.45 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr9_-_14314566 | 0.45 |
ENST00000397579.2
|
NFIB
|
nuclear factor I/B |
| chrX_+_135229731 | 0.44 |
ENST00000420362.1
|
FHL1
|
four and a half LIM domains 1 |
| chr10_+_104154229 | 0.44 |
ENST00000428099.1
ENST00000369966.3 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr2_+_74881398 | 0.44 |
ENST00000339773.5
ENST00000434486.1 |
SEMA4F
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
| chr11_+_64879317 | 0.44 |
ENST00000526809.1
ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2
|
transmembrane 7 superfamily member 2 |
| chr19_-_37407172 | 0.44 |
ENST00000391711.3
|
ZNF829
|
zinc finger protein 829 |
| chr19_+_40195101 | 0.43 |
ENST00000360675.3
ENST00000601802.1 |
LGALS14
|
lectin, galactoside-binding, soluble, 14 |
| chr19_+_10765699 | 0.43 |
ENST00000590009.1
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chrX_+_135229559 | 0.43 |
ENST00000394155.2
|
FHL1
|
four and a half LIM domains 1 |
| chr19_+_41117770 | 0.43 |
ENST00000601032.1
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr17_+_73089382 | 0.43 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr5_+_52285144 | 0.43 |
ENST00000296585.5
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
| chr4_-_78740511 | 0.43 |
ENST00000504123.1
ENST00000264903.4 ENST00000515441.1 |
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like |
| chr5_+_175299743 | 0.42 |
ENST00000502265.1
|
CPLX2
|
complexin 2 |
| chr3_-_183735731 | 0.42 |
ENST00000334444.6
|
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr3_+_154958715 | 0.41 |
ENST00000462531.1
ENST00000490497.1 |
RP11-451G4.1
|
RP11-451G4.1 |
| chr13_+_36050881 | 0.41 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr4_+_103423055 | 0.41 |
ENST00000505458.1
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr19_-_38743878 | 0.40 |
ENST00000587515.1
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr16_+_82068830 | 0.40 |
ENST00000199936.4
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr19_+_34287751 | 0.40 |
ENST00000590771.1
ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr4_+_144257915 | 0.39 |
ENST00000262995.4
|
GAB1
|
GRB2-associated binding protein 1 |
| chr7_-_38389573 | 0.39 |
ENST00000390344.2
|
TRGV5
|
T cell receptor gamma variable 5 |
| chr12_+_98909351 | 0.39 |
ENST00000343315.5
ENST00000266732.4 ENST00000393053.2 |
TMPO
|
thymopoietin |
| chr5_-_81046904 | 0.39 |
ENST00000515395.1
|
SSBP2
|
single-stranded DNA binding protein 2 |
| chr4_-_46996424 | 0.39 |
ENST00000264318.3
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4 |
| chr19_+_49404041 | 0.38 |
ENST00000263273.5
ENST00000424608.1 |
NUCB1
|
nucleobindin 1 |
| chr7_-_142247606 | 0.38 |
ENST00000390361.3
|
TRBV7-3
|
T cell receptor beta variable 7-3 |
| chr5_-_81046841 | 0.38 |
ENST00000509013.2
ENST00000505980.1 ENST00000509053.1 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr1_+_210406121 | 0.38 |
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4 |
| chr8_+_27348626 | 0.38 |
ENST00000517536.1
|
EPHX2
|
epoxide hydrolase 2, cytoplasmic |
| chr3_-_169381183 | 0.38 |
ENST00000494292.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr14_-_54955721 | 0.38 |
ENST00000554908.1
|
GMFB
|
glia maturation factor, beta |
| chrX_+_107069063 | 0.38 |
ENST00000262843.6
|
MID2
|
midline 2 |
| chr19_-_43586820 | 0.38 |
ENST00000406487.1
|
PSG2
|
pregnancy specific beta-1-glycoprotein 2 |
| chr12_-_23737534 | 0.37 |
ENST00000396007.2
|
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr2_+_56411131 | 0.37 |
ENST00000407595.2
|
CCDC85A
|
coiled-coil domain containing 85A |
| chr19_+_45844032 | 0.37 |
ENST00000589837.1
|
KLC3
|
kinesin light chain 3 |
| chr15_-_70390213 | 0.37 |
ENST00000557997.1
ENST00000317509.8 ENST00000442299.2 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr6_+_108881012 | 0.36 |
ENST00000343882.6
|
FOXO3
|
forkhead box O3 |
| chr12_-_96794143 | 0.36 |
ENST00000543119.2
|
CDK17
|
cyclin-dependent kinase 17 |
| chr7_+_127233689 | 0.36 |
ENST00000265825.5
ENST00000420086.2 |
FSCN3
|
fascin homolog 3, actin-bundling protein, testicular (Strongylocentrotus purpuratus) |
| chr12_-_6716569 | 0.36 |
ENST00000544040.1
ENST00000545942.1 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr12_-_10007448 | 0.36 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
Network of associatons between targets according to the STRING database.
First level regulatory network of EPAS1_BCL3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 1.2 | 6.0 | GO:1902228 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.8 | 2.4 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.8 | 2.3 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.6 | 1.7 | GO:1903248 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.5 | 1.6 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.5 | 1.4 | GO:0061216 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.4 | 2.6 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.4 | 1.3 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.4 | 1.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.4 | 1.1 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.3 | 1.4 | GO:0072255 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.3 | 1.6 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.3 | 0.8 | GO:0099545 | trans-synaptic signaling by trans-synaptic complex(GO:0099545) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.2 | 0.7 | GO:0055073 | cadmium ion homeostasis(GO:0055073) divalent metal ion export(GO:0070839) |
| 0.2 | 1.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.2 | 0.6 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.2 | 0.8 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.2 | 3.5 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 4.7 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 0.9 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.2 | 1.1 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.2 | 0.5 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.2 | 1.9 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.2 | 0.5 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.2 | 0.5 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.2 | 1.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 1.0 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.2 | 0.6 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.2 | 0.5 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.2 | 0.6 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 0.9 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.1 | 0.7 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.1 | 0.4 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.1 | 0.6 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 1.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.5 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.1 | 0.6 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.8 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.6 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 0.9 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 1.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.3 | GO:0002194 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.1 | 0.5 | GO:0009183 | ADP biosynthetic process(GO:0006172) purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.1 | 1.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.3 | GO:0045799 | mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 | 0.4 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.4 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 2.9 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 3.7 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 0.2 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.1 | 0.6 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.1 | 0.5 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 0.9 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.8 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.5 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 0.6 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.1 | 0.8 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.3 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.1 | 1.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.2 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.1 | 0.7 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.4 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.1 | 0.3 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.3 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 0.5 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.3 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 0.3 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.1 | 0.2 | GO:2000854 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) cellular response to cocaine(GO:0071314) positive regulation of corticosterone secretion(GO:2000854) |
| 0.1 | 0.1 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.1 | 0.9 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.1 | 0.5 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 0.1 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.1 | 0.3 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.3 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.3 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.4 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.1 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.2 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.1 | 0.6 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.2 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.4 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) response to aldosterone(GO:1904044) |
| 0.1 | 0.2 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.1 | 0.2 | GO:0060849 | radial pattern formation(GO:0009956) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 | 1.9 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.4 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.2 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.8 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 1.7 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.1 | 0.2 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 0.5 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.1 | 0.3 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.1 | 0.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.0 | 1.3 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.4 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.4 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.6 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.2 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.0 | 0.1 | GO:1905064 | negative regulation of vascular smooth muscle cell differentiation(GO:1905064) |
| 0.0 | 0.3 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.2 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.0 | 0.2 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.5 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.2 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.1 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.0 | 2.1 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 1.7 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 0.2 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.2 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 1.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.4 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.2 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.8 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.3 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.1 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.4 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.2 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.0 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.2 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.2 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.2 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.9 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.0 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.0 | 0.7 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.0 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.2 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.0 | 1.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.2 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.0 | 0.6 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.3 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.2 | GO:0032960 | regulation of inositol trisphosphate biosynthetic process(GO:0032960) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.6 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.2 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.9 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.1 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.0 | 0.2 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.5 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 0.2 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.9 | GO:1903859 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.1 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.1 | GO:0007442 | hindgut morphogenesis(GO:0007442) embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.8 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.4 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:1903570 | coronary vein morphogenesis(GO:0003169) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) cardiac vascular smooth muscle cell development(GO:0060948) positive regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061419) positive regulation of retinal ganglion cell axon guidance(GO:1902336) regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.9 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.3 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) |
| 0.0 | 0.4 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.4 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.1 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.5 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:1902219 | erythropoietin-mediated signaling pathway(GO:0038162) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.5 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.5 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.2 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.5 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.9 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.3 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.2 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.4 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 1.1 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.3 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.2 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:0060025 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) regulation of synaptic activity(GO:0060025) |
| 0.0 | 0.5 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.3 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.0 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.2 | GO:0099638 | endosome to plasma membrane protein transport(GO:0099638) |
| 0.0 | 0.4 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 1.9 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.0 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.5 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.2 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.4 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.2 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.1 | GO:0002329 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.7 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 1.0 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.0 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.2 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0030167 | proteoglycan catabolic process(GO:0030167) heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.0 | GO:0050779 | RNA destabilization(GO:0050779) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.6 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.1 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.3 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.1 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.3 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.0 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.3 | 1.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.3 | 2.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 2.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 1.2 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.2 | 3.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 0.5 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.2 | 0.5 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.4 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 0.7 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 1.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.9 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 2.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 1.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.3 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 1.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.4 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.7 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.3 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 0.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.5 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.3 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.1 | 0.4 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 0.2 | GO:0001534 | radial spoke(GO:0001534) |
| 0.1 | 0.8 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.2 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 3.2 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.2 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.1 | 0.2 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 1.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.5 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0005940 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.3 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 1.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.6 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 2.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.6 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.6 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 1.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) |
| 0.0 | 9.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 4.3 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.1 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.0 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.6 | 1.7 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.6 | 2.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.5 | 1.5 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.5 | 2.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.5 | 1.8 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.4 | 4.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 1.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.3 | 1.3 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.3 | 0.9 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.3 | 1.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.3 | 3.8 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 0.7 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.2 | 0.7 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.2 | 0.7 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.2 | 2.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 1.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 0.6 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.2 | 1.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.2 | 1.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 1.5 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.2 | 0.5 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.2 | 1.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 0.5 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.2 | 1.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.4 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 1.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 0.8 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 0.4 | GO:0032440 | 2-alkenal reductase [NAD(P)] activity(GO:0032440) |
| 0.1 | 0.8 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.1 | 2.9 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.5 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 3.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 1.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 2.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.4 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 0.5 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.6 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.7 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.6 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.5 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.3 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.1 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 1.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.2 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 0.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.3 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.1 | 0.3 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.1 | 1.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.2 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) leukemia inhibitory factor receptor activity(GO:0004923) interleukin-6 binding(GO:0019981) |
| 0.1 | 0.3 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.1 | 0.3 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.2 | GO:0052856 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.1 | 0.5 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.2 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.8 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.0 | 0.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 2.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.4 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 1.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.9 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.3 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.6 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.3 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 1.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.0 | 0.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 2.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 1.9 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.7 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0051990 | (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.0 | 0.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 1.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 1.6 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.2 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.0 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 1.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 3.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.6 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.1 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.0 | 0.0 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.3 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.0 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 1.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 1.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 3.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 4.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 5.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 3.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 2.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.5 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 1.8 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 1.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.7 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 2.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 3.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 5.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 2.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.8 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 2.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 1.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.2 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.6 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 2.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 3.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.6 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 1.6 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 2.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.5 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.4 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 1.1 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 2.0 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |