Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
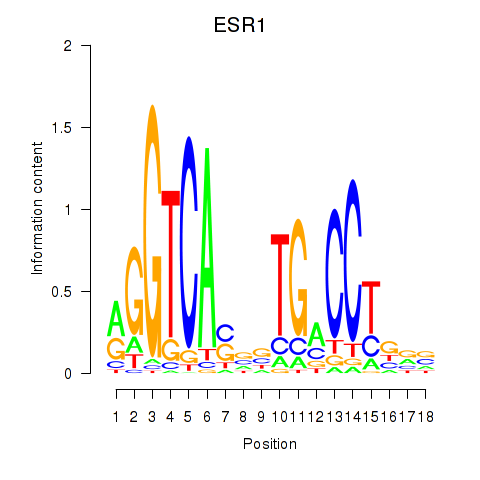
Results for ESR1
Z-value: 1.06
Transcription factors associated with ESR1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ESR1
|
ENSG00000091831.17 | estrogen receptor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ESR1 | hg19_v2_chr6_+_152128810_152128827 | 0.38 | 5.9e-02 | Click! |
Activity profile of ESR1 motif
Sorted Z-values of ESR1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_23046750 | 4.57 |
ENST00000390307.2
|
IGLV3-22
|
immunoglobulin lambda variable 3-22 (gene/pseudogene) |
| chr12_-_42877726 | 3.65 |
ENST00000548696.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr1_+_212458834 | 2.89 |
ENST00000261461.2
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr12_-_42877764 | 2.61 |
ENST00000455697.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr3_-_148804275 | 2.30 |
ENST00000392912.2
ENST00000465259.1 ENST00000310053.5 ENST00000494055.1 |
HLTF
|
helicase-like transcription factor |
| chr22_+_22936998 | 2.18 |
ENST00000390303.2
|
IGLV3-32
|
immunoglobulin lambda variable 3-32 (non-functional) |
| chr12_-_95044309 | 1.91 |
ENST00000261226.4
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr19_-_1513188 | 1.91 |
ENST00000330475.4
|
ADAMTSL5
|
ADAMTS-like 5 |
| chr3_+_38017264 | 1.85 |
ENST00000436654.1
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr3_-_122512619 | 1.84 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr20_-_45981138 | 1.83 |
ENST00000446994.2
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr3_+_37903432 | 1.81 |
ENST00000443503.2
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr9_-_134151915 | 1.77 |
ENST00000372271.3
|
FAM78A
|
family with sequence similarity 78, member A |
| chr16_-_88772670 | 1.75 |
ENST00000562544.1
|
RNF166
|
ring finger protein 166 |
| chr16_-_88772761 | 1.65 |
ENST00000567844.1
ENST00000312838.4 |
RNF166
|
ring finger protein 166 |
| chr19_-_18995029 | 1.52 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1 |
| chr13_+_103451548 | 1.52 |
ENST00000419638.1
|
BIVM
|
basic, immunoglobulin-like variable motif containing |
| chr8_-_28243590 | 1.52 |
ENST00000523095.1
ENST00000522795.1 |
ZNF395
|
zinc finger protein 395 |
| chr19_+_58790314 | 1.51 |
ENST00000196548.5
ENST00000608843.1 |
ZNF8
ZNF8
|
Zinc finger protein 8 zinc finger protein 8 |
| chr13_+_103451399 | 1.47 |
ENST00000257336.1
ENST00000448849.2 |
BIVM
|
basic, immunoglobulin-like variable motif containing |
| chr11_+_47270436 | 1.47 |
ENST00000395397.3
ENST00000405576.1 |
NR1H3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr11_+_47270475 | 1.44 |
ENST00000481889.2
ENST00000436778.1 ENST00000531660.1 ENST00000407404.1 |
NR1H3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr4_-_89080003 | 1.43 |
ENST00000237612.3
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr19_+_50194360 | 1.42 |
ENST00000323446.5
ENST00000392518.4 ENST00000598396.1 ENST00000598293.1 ENST00000354199.5 ENST00000405931.2 ENST00000602019.1 |
CPT1C
|
carnitine palmitoyltransferase 1C |
| chr8_-_28243934 | 1.42 |
ENST00000521185.1
ENST00000520290.1 ENST00000344423.5 |
ZNF395
|
zinc finger protein 395 |
| chr2_-_86094764 | 1.42 |
ENST00000393808.3
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr2_+_233925064 | 1.37 |
ENST00000359570.5
ENST00000538935.1 |
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr13_-_103451307 | 1.37 |
ENST00000376004.4
|
KDELC1
|
KDEL (Lys-Asp-Glu-Leu) containing 1 |
| chr14_-_106453155 | 1.35 |
ENST00000390594.2
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr3_+_58223228 | 1.31 |
ENST00000478253.1
ENST00000295962.4 |
ABHD6
|
abhydrolase domain containing 6 |
| chr11_-_118134997 | 1.31 |
ENST00000278937.2
|
MPZL2
|
myelin protein zero-like 2 |
| chr17_-_46688334 | 1.29 |
ENST00000239165.7
|
HOXB7
|
homeobox B7 |
| chr2_-_132559234 | 1.26 |
ENST00000303798.2
|
C2orf27B
|
chromosome 2 open reading frame 27B |
| chr4_-_99578776 | 1.24 |
ENST00000515287.1
|
TSPAN5
|
tetraspanin 5 |
| chr19_-_13213662 | 1.21 |
ENST00000264824.4
|
LYL1
|
lymphoblastic leukemia derived sequence 1 |
| chr1_+_901847 | 1.15 |
ENST00000379410.3
ENST00000379409.2 ENST00000379407.3 |
PLEKHN1
|
pleckstrin homology domain containing, family N member 1 |
| chr10_+_54074033 | 1.10 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr16_-_75300679 | 1.08 |
ENST00000420641.3
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr22_+_32455111 | 1.06 |
ENST00000543737.1
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr4_-_99578789 | 1.06 |
ENST00000511651.1
ENST00000505184.1 |
TSPAN5
|
tetraspanin 5 |
| chr10_-_7513904 | 1.03 |
ENST00000420395.1
|
RP5-1031D4.2
|
RP5-1031D4.2 |
| chr4_-_140201333 | 1.02 |
ENST00000398955.1
|
MGARP
|
mitochondria-localized glutamic acid-rich protein |
| chr9_+_140172200 | 1.02 |
ENST00000357503.2
|
TOR4A
|
torsin family 4, member A |
| chr3_-_183735651 | 1.01 |
ENST00000427120.2
ENST00000392579.2 ENST00000382494.2 ENST00000446941.2 |
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr1_-_16763685 | 0.99 |
ENST00000540400.1
|
SPATA21
|
spermatogenesis associated 21 |
| chr6_+_32121789 | 0.93 |
ENST00000437001.2
ENST00000375137.2 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr6_+_24495067 | 0.92 |
ENST00000357578.3
ENST00000546278.1 ENST00000491546.1 |
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr6_+_32121908 | 0.90 |
ENST00000375143.2
ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr14_-_105420241 | 0.90 |
ENST00000557457.1
|
AHNAK2
|
AHNAK nucleoprotein 2 |
| chr14_+_89290965 | 0.88 |
ENST00000345383.5
ENST00000536576.1 ENST00000346301.4 ENST00000338104.6 ENST00000354441.6 ENST00000380656.2 ENST00000556651.1 ENST00000554686.1 |
TTC8
|
tetratricopeptide repeat domain 8 |
| chr2_+_191208196 | 0.86 |
ENST00000392329.2
ENST00000322522.4 ENST00000430311.1 ENST00000541441.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chrX_+_77166172 | 0.86 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr20_+_1246908 | 0.86 |
ENST00000381873.3
ENST00000381867.1 |
SNPH
|
syntaphilin |
| chr22_+_29702996 | 0.86 |
ENST00000406549.3
ENST00000360113.2 ENST00000341313.6 ENST00000403764.1 ENST00000471961.1 ENST00000407854.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chr1_+_16083154 | 0.83 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr6_+_24495185 | 0.83 |
ENST00000348925.2
|
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr19_+_7011509 | 0.83 |
ENST00000377296.3
|
AC025278.1
|
Uncharacterized protein |
| chr1_-_59043166 | 0.82 |
ENST00000371225.2
|
TACSTD2
|
tumor-associated calcium signal transducer 2 |
| chr7_-_132261253 | 0.82 |
ENST00000321063.4
|
PLXNA4
|
plexin A4 |
| chr19_-_49176264 | 0.81 |
ENST00000270235.4
ENST00000596844.1 |
NTN5
|
netrin 5 |
| chr16_+_89894875 | 0.81 |
ENST00000393062.2
|
SPIRE2
|
spire-type actin nucleation factor 2 |
| chr17_-_39677971 | 0.80 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr3_-_183735731 | 0.80 |
ENST00000334444.6
|
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr17_+_37793318 | 0.79 |
ENST00000336308.5
|
STARD3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr6_-_2971429 | 0.79 |
ENST00000380529.1
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr19_-_19051993 | 0.77 |
ENST00000594794.1
ENST00000355887.6 ENST00000392351.3 ENST00000596482.1 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr14_-_106066376 | 0.76 |
ENST00000412518.1
ENST00000428654.1 ENST00000427543.1 ENST00000420153.1 ENST00000577108.1 ENST00000576077.1 |
AL928742.12
IGHE
|
AL928742.12 immunoglobulin heavy constant epsilon |
| chr6_+_32121218 | 0.76 |
ENST00000414204.1
ENST00000361568.2 ENST00000395523.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr14_-_75536182 | 0.76 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr11_+_33563821 | 0.75 |
ENST00000321505.4
ENST00000265654.5 ENST00000389726.3 |
KIAA1549L
|
KIAA1549-like |
| chr6_-_2971494 | 0.74 |
ENST00000380539.1
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr13_+_37006398 | 0.74 |
ENST00000418263.1
|
CCNA1
|
cyclin A1 |
| chr22_-_37880543 | 0.73 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr19_-_17375527 | 0.72 |
ENST00000431146.2
ENST00000594190.1 |
USHBP1
|
Usher syndrome 1C binding protein 1 |
| chr7_-_16685422 | 0.72 |
ENST00000306999.2
|
ANKMY2
|
ankyrin repeat and MYND domain containing 2 |
| chr5_+_110559784 | 0.72 |
ENST00000282356.4
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr11_-_47270341 | 0.71 |
ENST00000529444.1
ENST00000530453.1 ENST00000537863.1 ENST00000529788.1 ENST00000444355.2 ENST00000527256.1 ENST00000529663.1 ENST00000256997.3 |
ACP2
|
acid phosphatase 2, lysosomal |
| chr12_-_14996355 | 0.71 |
ENST00000228936.4
|
ART4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr6_-_111804905 | 0.71 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr22_+_23165153 | 0.71 |
ENST00000390317.2
|
IGLV2-8
|
immunoglobulin lambda variable 2-8 |
| chr11_+_3876859 | 0.70 |
ENST00000300737.4
|
STIM1
|
stromal interaction molecule 1 |
| chr4_-_10023095 | 0.69 |
ENST00000264784.3
|
SLC2A9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr6_+_139094657 | 0.69 |
ENST00000332797.6
|
CCDC28A
|
coiled-coil domain containing 28A |
| chr17_+_37793378 | 0.69 |
ENST00000544210.2
ENST00000581894.1 ENST00000394250.4 ENST00000579479.1 ENST00000577248.1 ENST00000580611.1 |
STARD3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr7_+_94139105 | 0.68 |
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1 |
| chr9_+_128509663 | 0.67 |
ENST00000373489.5
ENST00000373483.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr5_+_170288856 | 0.66 |
ENST00000523189.1
|
RANBP17
|
RAN binding protein 17 |
| chr19_+_46002868 | 0.66 |
ENST00000396735.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr12_+_125478241 | 0.66 |
ENST00000341446.8
|
BRI3BP
|
BRI3 binding protein |
| chr3_-_46506358 | 0.65 |
ENST00000417439.1
ENST00000431944.1 |
LTF
|
lactotransferrin |
| chr10_+_81466084 | 0.64 |
ENST00000342531.2
|
NUTM2B
|
NUT family member 2B |
| chr14_-_67955426 | 0.64 |
ENST00000554480.1
|
TMEM229B
|
transmembrane protein 229B |
| chr17_-_19771242 | 0.64 |
ENST00000361658.2
|
ULK2
|
unc-51 like autophagy activating kinase 2 |
| chr11_+_64107663 | 0.64 |
ENST00000356786.5
|
CCDC88B
|
coiled-coil domain containing 88B |
| chr3_+_48488114 | 0.62 |
ENST00000421175.1
ENST00000320211.3 ENST00000346691.4 ENST00000357105.6 |
ATRIP
|
ATR interacting protein |
| chr13_+_37006421 | 0.62 |
ENST00000255465.4
|
CCNA1
|
cyclin A1 |
| chr11_+_123396528 | 0.62 |
ENST00000322282.7
ENST00000529750.1 |
GRAMD1B
|
GRAM domain containing 1B |
| chr3_-_128294929 | 0.62 |
ENST00000356020.2
|
C3orf27
|
chromosome 3 open reading frame 27 |
| chr1_+_43291220 | 0.62 |
ENST00000372514.3
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group) |
| chr1_+_245133656 | 0.62 |
ENST00000366521.3
|
EFCAB2
|
EF-hand calcium binding domain 2 |
| chr19_-_58864848 | 0.62 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr11_+_57310114 | 0.60 |
ENST00000527972.1
ENST00000399154.2 |
SMTNL1
|
smoothelin-like 1 |
| chr22_-_51016433 | 0.60 |
ENST00000405237.3
|
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr13_+_37005967 | 0.58 |
ENST00000440264.1
ENST00000449823.1 |
CCNA1
|
cyclin A1 |
| chr6_-_2903514 | 0.58 |
ENST00000380698.4
|
SERPINB9
|
serpin peptidase inhibitor, clade B (ovalbumin), member 9 |
| chr1_+_33231268 | 0.58 |
ENST00000373480.1
|
KIAA1522
|
KIAA1522 |
| chr6_+_24775153 | 0.57 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr10_-_103880209 | 0.57 |
ENST00000425280.1
|
LDB1
|
LIM domain binding 1 |
| chr3_+_42897512 | 0.57 |
ENST00000493193.1
|
ACKR2
|
atypical chemokine receptor 2 |
| chr11_+_62439126 | 0.57 |
ENST00000377953.3
|
C11orf83
|
chromosome 11 open reading frame 83 |
| chr11_-_66445219 | 0.56 |
ENST00000525754.1
ENST00000531969.1 ENST00000524637.1 ENST00000531036.2 ENST00000310046.4 |
RBM4B
|
RNA binding motif protein 4B |
| chr6_-_32122106 | 0.56 |
ENST00000428778.1
|
PRRT1
|
proline-rich transmembrane protein 1 |
| chr14_-_105531759 | 0.56 |
ENST00000329797.3
ENST00000539291.2 ENST00000392585.2 |
GPR132
|
G protein-coupled receptor 132 |
| chr7_+_149535455 | 0.56 |
ENST00000223210.4
ENST00000460379.1 |
ZNF862
|
zinc finger protein 862 |
| chr8_-_71316021 | 0.55 |
ENST00000452400.2
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr22_+_31277661 | 0.54 |
ENST00000454145.1
ENST00000453621.1 ENST00000431368.1 ENST00000535268.1 |
OSBP2
|
oxysterol binding protein 2 |
| chr2_+_177015122 | 0.54 |
ENST00000468418.3
|
HOXD3
|
homeobox D3 |
| chr4_-_101111615 | 0.54 |
ENST00000273990.2
|
DDIT4L
|
DNA-damage-inducible transcript 4-like |
| chr19_-_17375541 | 0.53 |
ENST00000252597.3
|
USHBP1
|
Usher syndrome 1C binding protein 1 |
| chr11_-_63376013 | 0.53 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr7_-_99766191 | 0.52 |
ENST00000423751.1
ENST00000360039.4 |
GAL3ST4
|
galactose-3-O-sulfotransferase 4 |
| chrX_-_148676974 | 0.52 |
ENST00000524178.1
|
HSFX2
|
heat shock transcription factor family, X linked 2 |
| chr16_-_10652993 | 0.52 |
ENST00000536829.1
|
EMP2
|
epithelial membrane protein 2 |
| chr3_-_33759699 | 0.51 |
ENST00000399362.4
ENST00000359576.5 ENST00000307312.7 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr8_-_101571964 | 0.51 |
ENST00000520552.1
ENST00000521345.1 ENST00000523000.1 ENST00000335659.3 ENST00000358990.3 ENST00000519597.1 |
ANKRD46
|
ankyrin repeat domain 46 |
| chr2_+_182756615 | 0.51 |
ENST00000431877.2
ENST00000320370.7 |
SSFA2
|
sperm specific antigen 2 |
| chr2_+_242750274 | 0.51 |
ENST00000405370.1
|
NEU4
|
sialidase 4 |
| chr17_+_31254892 | 0.51 |
ENST00000394642.3
ENST00000579849.1 |
TMEM98
|
transmembrane protein 98 |
| chr1_-_204121102 | 0.50 |
ENST00000367202.4
|
ETNK2
|
ethanolamine kinase 2 |
| chr17_-_76274572 | 0.50 |
ENST00000374945.1
|
RP11-219G17.4
|
RP11-219G17.4 |
| chr1_-_40137710 | 0.50 |
ENST00000235628.1
|
NT5C1A
|
5'-nucleotidase, cytosolic IA |
| chrX_+_148855726 | 0.50 |
ENST00000370416.4
|
HSFX1
|
heat shock transcription factor family, X linked 1 |
| chr4_+_25915822 | 0.49 |
ENST00000506197.2
|
SMIM20
|
small integral membrane protein 20 |
| chr5_-_73936451 | 0.49 |
ENST00000537006.1
|
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr11_+_65190245 | 0.48 |
ENST00000499732.1
ENST00000501122.2 ENST00000601801.1 |
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr3_-_33759541 | 0.48 |
ENST00000468888.2
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr6_+_28092338 | 0.48 |
ENST00000340487.4
|
ZSCAN16
|
zinc finger and SCAN domain containing 16 |
| chr9_+_141107506 | 0.48 |
ENST00000446912.2
|
FAM157B
|
family with sequence similarity 157, member B |
| chr8_-_41522719 | 0.48 |
ENST00000335651.6
|
ANK1
|
ankyrin 1, erythrocytic |
| chr12_+_112204691 | 0.48 |
ENST00000416293.3
ENST00000261733.2 |
ALDH2
|
aldehyde dehydrogenase 2 family (mitochondrial) |
| chr10_+_74870253 | 0.48 |
ENST00000544879.1
ENST00000537969.1 ENST00000372997.3 |
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr8_-_145690724 | 0.48 |
ENST00000526887.1
ENST00000533764.1 ENST00000403000.2 |
CYHR1
|
cysteine/histidine-rich 1 |
| chr9_-_127263265 | 0.47 |
ENST00000373587.3
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr7_+_155090271 | 0.47 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr1_-_24151892 | 0.47 |
ENST00000235958.4
|
HMGCL
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr1_-_24151903 | 0.47 |
ENST00000436439.2
ENST00000374490.3 |
HMGCL
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr14_-_91884150 | 0.46 |
ENST00000553403.1
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr12_+_123874589 | 0.46 |
ENST00000437502.1
|
SETD8
|
SET domain containing (lysine methyltransferase) 8 |
| chr13_-_99174252 | 0.46 |
ENST00000376547.3
|
STK24
|
serine/threonine kinase 24 |
| chr19_-_2740036 | 0.46 |
ENST00000586572.1
ENST00000269740.4 |
AC006538.4
SLC39A3
|
Uncharacterized protein solute carrier family 39 (zinc transporter), member 3 |
| chr17_-_41277467 | 0.46 |
ENST00000494123.1
ENST00000346315.3 ENST00000309486.4 ENST00000468300.1 ENST00000354071.3 ENST00000352993.3 ENST00000471181.2 |
BRCA1
|
breast cancer 1, early onset |
| chr17_-_34592032 | 0.46 |
ENST00000457979.3
|
TBC1D3C
|
TBC1 domain family, member 3C |
| chr14_+_105992906 | 0.45 |
ENST00000392519.2
|
TMEM121
|
transmembrane protein 121 |
| chr2_+_219264466 | 0.45 |
ENST00000273062.2
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr14_-_106054659 | 0.45 |
ENST00000390539.2
|
IGHA2
|
immunoglobulin heavy constant alpha 2 (A2m marker) |
| chr10_+_76586348 | 0.45 |
ENST00000372724.1
ENST00000287239.4 ENST00000372714.1 |
KAT6B
|
K(lysine) acetyltransferase 6B |
| chr1_+_94883991 | 0.44 |
ENST00000370214.4
|
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr7_-_134001663 | 0.44 |
ENST00000378509.4
|
SLC35B4
|
solute carrier family 35 (UDP-xylose/UDP-N-acetylglucosamine transporter), member B4 |
| chr12_-_52585765 | 0.44 |
ENST00000313234.5
ENST00000394815.2 |
KRT80
|
keratin 80 |
| chr3_-_125775629 | 0.44 |
ENST00000383598.2
|
SLC41A3
|
solute carrier family 41, member 3 |
| chr5_+_32710736 | 0.44 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr3_-_93781750 | 0.44 |
ENST00000314636.2
|
DHFRL1
|
dihydrofolate reductase-like 1 |
| chr17_+_43922241 | 0.43 |
ENST00000329196.5
|
SPPL2C
|
signal peptide peptidase like 2C |
| chr10_+_74870206 | 0.43 |
ENST00000357321.4
ENST00000349051.5 |
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr1_-_16763911 | 0.43 |
ENST00000375577.1
ENST00000335496.1 |
SPATA21
|
spermatogenesis associated 21 |
| chr8_+_42010464 | 0.43 |
ENST00000518421.1
ENST00000174653.3 ENST00000396926.3 ENST00000521280.1 ENST00000522288.1 |
AP3M2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr17_+_36283971 | 0.42 |
ENST00000327454.6
ENST00000378174.5 |
TBC1D3F
|
TBC1 domain family, member 3F |
| chr22_-_21579843 | 0.42 |
ENST00000405188.4
|
GGT2
|
gamma-glutamyltransferase 2 |
| chr17_-_27507377 | 0.42 |
ENST00000531253.1
|
MYO18A
|
myosin XVIIIA |
| chr16_-_3306587 | 0.42 |
ENST00000541159.1
ENST00000536379.1 ENST00000219596.1 ENST00000339854.4 |
MEFV
|
Mediterranean fever |
| chr17_-_34757039 | 0.42 |
ENST00000455054.2
ENST00000308078.7 |
TBC1D3H
TBC1D3C
|
TBC1 domain family, member 3H TBC1 domain family, member 3C |
| chr1_-_27286897 | 0.42 |
ENST00000320567.5
|
C1orf172
|
chromosome 1 open reading frame 172 |
| chr17_-_36348610 | 0.42 |
ENST00000339023.4
ENST00000354664.4 |
TBC1D3
|
TBC1 domain family, member 3 |
| chr7_-_30544405 | 0.42 |
ENST00000409390.1
ENST00000409144.1 ENST00000005374.6 ENST00000409436.1 ENST00000275428.4 |
GGCT
|
gamma-glutamylcyclotransferase |
| chr15_-_20170354 | 0.42 |
ENST00000338912.5
|
IGHV1OR15-9
|
immunoglobulin heavy variable 1/OR15-9 (non-functional) |
| chr17_-_27507395 | 0.42 |
ENST00000354329.4
ENST00000527372.1 |
MYO18A
|
myosin XVIIIA |
| chr17_-_34808047 | 0.42 |
ENST00000592614.1
ENST00000591542.1 ENST00000330458.7 ENST00000341264.6 ENST00000592987.1 ENST00000400684.4 |
TBC1D3G
TBC1D3H
|
TBC1 domain family, member 3G TBC1 domain family, member 3H |
| chr1_-_98386543 | 0.42 |
ENST00000423006.2
ENST00000370192.3 ENST00000306031.5 |
DPYD
|
dihydropyrimidine dehydrogenase |
| chr5_+_175288631 | 0.42 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr10_+_112257596 | 0.41 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5 |
| chr14_-_100841930 | 0.41 |
ENST00000555031.1
ENST00000553395.1 ENST00000553545.1 ENST00000344102.5 ENST00000556338.1 ENST00000392882.2 ENST00000553934.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr11_-_1780261 | 0.41 |
ENST00000427721.1
|
RP11-295K3.1
|
RP11-295K3.1 |
| chr19_+_42829702 | 0.41 |
ENST00000334370.4
|
MEGF8
|
multiple EGF-like-domains 8 |
| chr2_-_219696519 | 0.41 |
ENST00000545803.1
ENST00000430489.1 ENST00000392098.3 |
PRKAG3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit |
| chr19_-_19843900 | 0.40 |
ENST00000344099.3
|
ZNF14
|
zinc finger protein 14 |
| chr4_+_110481348 | 0.40 |
ENST00000394650.4
|
CCDC109B
|
coiled-coil domain containing 109B |
| chr22_+_39745930 | 0.40 |
ENST00000318801.4
ENST00000216155.7 ENST00000406293.3 ENST00000328933.5 |
SYNGR1
|
synaptogyrin 1 |
| chr7_+_2727817 | 0.40 |
ENST00000407112.1
|
AMZ1
|
archaelysin family metallopeptidase 1 |
| chr4_+_25915896 | 0.40 |
ENST00000514384.1
|
SMIM20
|
small integral membrane protein 20 |
| chr6_-_41006928 | 0.40 |
ENST00000244565.3
|
UNC5CL
|
unc-5 homolog C (C. elegans)-like |
| chr10_+_120863587 | 0.40 |
ENST00000535029.1
ENST00000361432.2 ENST00000544016.1 |
FAM45A
|
family with sequence similarity 45, member A |
| chr18_-_11908329 | 0.40 |
ENST00000344987.7
ENST00000588103.1 ENST00000588191.1 ENST00000317235.7 ENST00000309976.9 ENST00000588186.1 ENST00000589267.1 |
MPPE1
|
metallophosphoesterase 1 |
| chr3_+_169684553 | 0.40 |
ENST00000337002.4
ENST00000480708.1 |
SEC62
|
SEC62 homolog (S. cerevisiae) |
| chr4_-_130692631 | 0.40 |
ENST00000500092.2
ENST00000509105.1 |
RP11-519M16.1
|
RP11-519M16.1 |
| chr11_+_66624527 | 0.40 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr2_-_201828356 | 0.39 |
ENST00000234296.2
|
ORC2
|
origin recognition complex, subunit 2 |
| chr6_+_33378738 | 0.39 |
ENST00000374512.3
ENST00000374516.3 |
PHF1
|
PHD finger protein 1 |
| chr5_+_57878859 | 0.39 |
ENST00000282878.4
|
RAB3C
|
RAB3C, member RAS oncogene family |
| chr3_-_138313161 | 0.39 |
ENST00000489254.1
ENST00000474781.1 ENST00000462419.1 ENST00000264982.3 |
CEP70
|
centrosomal protein 70kDa |
| chr7_+_101460882 | 0.38 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ESR1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:2000691 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 1.0 | 2.9 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.5 | 1.5 | GO:0036146 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.5 | 2.9 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.5 | 1.4 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.4 | 1.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.4 | 1.1 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.3 | 1.8 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.3 | 1.8 | GO:0009450 | acetate metabolic process(GO:0006083) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.3 | 0.9 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.3 | 0.8 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.3 | 2.6 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 0.7 | GO:0033214 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 0.2 | 1.0 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.2 | 0.6 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.2 | 1.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 0.7 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 0.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 0.6 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.2 | 0.5 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 1.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.2 | 0.5 | GO:0070512 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.1 | 1.5 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.1 | 0.4 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 1.2 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.1 | 0.8 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.5 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.5 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) |
| 0.1 | 1.3 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 1.4 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 1.0 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.1 | 0.5 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 0.8 | GO:0070649 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 0.7 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 1.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.9 | GO:0050893 | sensory processing(GO:0050893) |
| 0.1 | 0.3 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.1 | 0.5 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 1.8 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.4 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.1 | 0.9 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 0.4 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 | 0.7 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.7 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.1 | 0.3 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.1 | 0.6 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.1 | 0.6 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.1 | 0.6 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 0.3 | GO:0071623 | complement component C5a signaling pathway(GO:0038178) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.3 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 | 0.3 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.1 | 0.5 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.3 | GO:0042853 | L-alanine metabolic process(GO:0042851) L-alanine catabolic process(GO:0042853) |
| 0.1 | 0.2 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.1 | 1.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 1.0 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 1.0 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.7 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.1 | 0.6 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.3 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.2 | GO:0060931 | sinoatrial node development(GO:0003163) sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 0.1 | 0.9 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 1.9 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.2 | GO:1903625 | L-ascorbic acid biosynthetic process(GO:0019853) negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.1 | 0.7 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.2 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.1 | 0.2 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 0.1 | 1.3 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.1 | 0.8 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.5 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.1 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.2 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.4 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.1 | 0.5 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 1.3 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 0.3 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.1 | 0.2 | GO:1903215 | regulation of kidney size(GO:0035564) positive regulation of protein autoubiquitination(GO:1902499) negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.1 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.0 | 0.2 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.0 | 0.3 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.0 | 0.4 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.2 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.4 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.0 | 0.3 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.3 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.9 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.5 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 0.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.2 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.8 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 2.8 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.3 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.4 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.4 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.2 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.3 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.3 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.2 | GO:1901091 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.0 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 1.1 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.3 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.4 | GO:2000781 | DNA dealkylation involved in DNA repair(GO:0006307) positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 0.3 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 1.5 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.6 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.4 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 2.8 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 3.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.3 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.3 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.7 | GO:0006525 | arginine metabolic process(GO:0006525) |
| 0.0 | 0.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 3.8 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.3 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.4 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 1.4 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.6 | GO:0036344 | platelet morphogenesis(GO:0036344) |
| 0.0 | 0.0 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.0 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.4 | GO:0033866 | nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 3.6 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 2.4 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.2 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:1902951 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.4 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.4 | GO:0061082 | myeloid leukocyte cytokine production(GO:0061082) |
| 0.0 | 0.2 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 1.0 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.3 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.2 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.1 | GO:1901569 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.0 | 0.3 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.3 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.4 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.0 | 0.1 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.3 | GO:0043651 | alpha-linolenic acid metabolic process(GO:0036109) linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.1 | GO:0045991 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) carbon catabolite activation of transcription(GO:0045991) positive regulation of transcription by glucose(GO:0046016) |
| 0.0 | 0.5 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.3 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.2 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.4 | GO:0043536 | positive regulation of blood vessel endothelial cell migration(GO:0043536) |
| 0.0 | 0.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.2 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.8 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 1.5 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.4 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.1 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.0 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 1.5 | GO:0006664 | glycolipid metabolic process(GO:0006664) liposaccharide metabolic process(GO:1903509) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.1 | 0.4 | GO:0071751 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.1 | 0.4 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.1 | 0.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.7 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.5 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.1 | 0.7 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 2.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.7 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.4 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.1 | 0.7 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 1.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 1.0 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.9 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.2 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 2.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 1.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 1.0 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 1.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.2 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.0 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 2.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 1.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 3.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.9 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.9 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.5 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 2.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.2 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.3 | 2.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.3 | 0.9 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.3 | 2.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 1.4 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.2 | 0.9 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.2 | 0.7 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 1.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.4 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 0.4 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.1 | 0.4 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.5 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.5 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.1 | 1.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.8 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.4 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.1 | 1.6 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 0.3 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.6 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 0.5 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 1.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.7 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.3 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 0.3 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.9 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.3 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.1 | 1.4 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.1 | 0.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 1.0 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.9 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 0.2 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.1 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.2 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.1 | 0.7 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 1.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.5 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 1.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 1.4 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 0.3 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.1 | 3.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.2 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 1.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.6 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 0.4 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.1 | 0.2 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.1 | 0.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.5 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.3 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 1.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.6 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 1.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 2.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 1.0 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.2 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 6.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 1.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.2 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.1 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.7 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.4 | GO:0015216 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.4 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 1.7 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.1 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.9 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.6 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 2.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.3 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 2.2 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 3.7 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 3.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 2.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 3.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.8 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 2.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.7 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 1.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 1.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 0.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 0.7 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 0.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 1.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 3.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |