Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
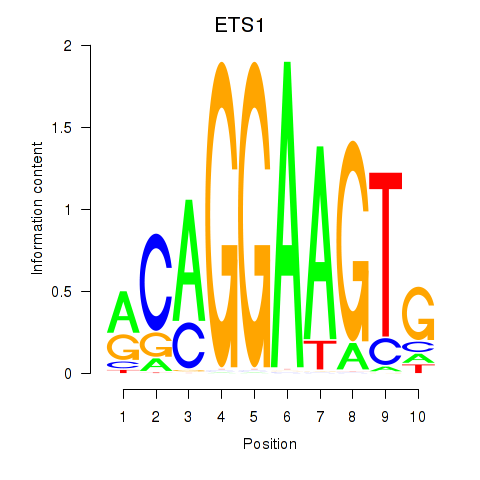
Results for ETS1
Z-value: 1.06
Transcription factors associated with ETS1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETS1
|
ENSG00000134954.10 | ETS proto-oncogene 1, transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ETS1 | hg19_v2_chr11_-_128457446_128457461 | -0.46 | 2.1e-02 | Click! |
Activity profile of ETS1 motif
Sorted Z-values of ETS1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_225266743 | 4.52 |
ENST00000409685.3
|
FAM124B
|
family with sequence similarity 124B |
| chr2_-_225266711 | 4.50 |
ENST00000389874.3
|
FAM124B
|
family with sequence similarity 124B |
| chr9_-_134145880 | 4.50 |
ENST00000372269.3
ENST00000464831.1 |
FAM78A
|
family with sequence similarity 78, member A |
| chr1_+_35258592 | 4.48 |
ENST00000342280.4
ENST00000450137.1 |
GJA4
|
gap junction protein, alpha 4, 37kDa |
| chr12_+_53443963 | 4.36 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr12_+_53443680 | 4.35 |
ENST00000314250.6
ENST00000451358.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr2_+_31456874 | 3.51 |
ENST00000541626.1
|
EHD3
|
EH-domain containing 3 |
| chr3_-_18480260 | 3.39 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr20_-_23030296 | 3.08 |
ENST00000377103.2
|
THBD
|
thrombomodulin |
| chr4_-_16900410 | 3.06 |
ENST00000304523.5
|
LDB2
|
LIM domain binding 2 |
| chr4_-_16900184 | 3.05 |
ENST00000515064.1
|
LDB2
|
LIM domain binding 2 |
| chr4_-_16900242 | 3.02 |
ENST00000502640.1
ENST00000506732.1 |
LDB2
|
LIM domain binding 2 |
| chr4_-_16900217 | 3.01 |
ENST00000441778.2
|
LDB2
|
LIM domain binding 2 |
| chr1_+_84609944 | 2.96 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr16_-_67260691 | 2.81 |
ENST00000447579.1
ENST00000393992.1 ENST00000424285.1 |
LRRC29
|
leucine rich repeat containing 29 |
| chr4_-_101439242 | 2.81 |
ENST00000296420.4
|
EMCN
|
endomucin |
| chr6_+_142622991 | 2.81 |
ENST00000230173.6
ENST00000367608.2 |
GPR126
|
G protein-coupled receptor 126 |
| chr11_-_118122996 | 2.76 |
ENST00000525386.1
ENST00000527472.1 ENST00000278949.4 |
MPZL3
|
myelin protein zero-like 3 |
| chr14_-_54421190 | 2.69 |
ENST00000417573.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr14_-_38725573 | 2.58 |
ENST00000342213.2
|
CLEC14A
|
C-type lectin domain family 14, member A |
| chr1_+_84543734 | 2.54 |
ENST00000370689.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr9_-_77703115 | 2.50 |
ENST00000361092.4
ENST00000376808.4 |
NMRK1
|
nicotinamide riboside kinase 1 |
| chr9_-_77703056 | 2.49 |
ENST00000376811.1
|
NMRK1
|
nicotinamide riboside kinase 1 |
| chr4_-_186696425 | 2.39 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_-_69870747 | 2.38 |
ENST00000409068.1
|
AAK1
|
AP2 associated kinase 1 |
| chr1_+_61547894 | 2.33 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr9_-_134151915 | 2.27 |
ENST00000372271.3
|
FAM78A
|
family with sequence similarity 78, member A |
| chr4_-_101439148 | 2.27 |
ENST00000511970.1
ENST00000502569.1 ENST00000305864.3 |
EMCN
|
endomucin |
| chrX_-_125686784 | 2.23 |
ENST00000371126.1
|
DCAF12L1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr10_+_94590910 | 2.20 |
ENST00000371547.4
|
EXOC6
|
exocyst complex component 6 |
| chr4_-_186733363 | 2.19 |
ENST00000393523.2
ENST00000393528.3 ENST00000449407.2 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_+_177134201 | 2.17 |
ENST00000452865.1
|
MTX2
|
metaxin 2 |
| chr1_+_61548225 | 2.16 |
ENST00000371187.3
|
NFIA
|
nuclear factor I/A |
| chr8_-_22550815 | 2.16 |
ENST00000317216.2
|
EGR3
|
early growth response 3 |
| chr16_+_4845379 | 2.14 |
ENST00000588606.1
ENST00000586005.1 |
SMIM22
|
small integral membrane protein 22 |
| chr7_-_150329421 | 2.09 |
ENST00000493969.1
ENST00000328902.5 |
GIMAP6
|
GTPase, IMAP family member 6 |
| chr8_+_22438009 | 1.97 |
ENST00000409417.1
|
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr2_+_177134134 | 1.97 |
ENST00000249442.6
ENST00000392529.2 ENST00000443241.1 |
MTX2
|
metaxin 2 |
| chr5_+_61874562 | 1.96 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr7_+_95401851 | 1.96 |
ENST00000447467.2
|
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chrX_+_77166172 | 1.95 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr17_+_40913264 | 1.92 |
ENST00000587142.1
ENST00000588576.1 |
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr16_-_67260901 | 1.89 |
ENST00000341546.3
ENST00000409509.1 ENST00000433915.1 ENST00000454102.2 |
LRRC29
AC040160.1
|
leucine rich repeat containing 29 Uncharacterized protein; cDNA FLJ57407, weakly similar to Mus musculus leucine rich repeat containing 29 (Lrrc29), mRNA |
| chr7_+_150264365 | 1.89 |
ENST00000255945.2
ENST00000461940.1 |
GIMAP4
|
GTPase, IMAP family member 4 |
| chr9_-_130617029 | 1.87 |
ENST00000373203.4
|
ENG
|
endoglin |
| chr1_+_109102652 | 1.86 |
ENST00000370035.3
ENST00000405454.1 |
FAM102B
|
family with sequence similarity 102, member B |
| chr11_+_59522900 | 1.86 |
ENST00000529177.1
|
STX3
|
syntaxin 3 |
| chr8_+_29952914 | 1.85 |
ENST00000321250.8
ENST00000518001.1 ENST00000520682.1 ENST00000442880.2 ENST00000523116.1 |
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chr12_+_9067123 | 1.85 |
ENST00000543824.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr9_-_130616915 | 1.83 |
ENST00000344849.3
|
ENG
|
endoglin |
| chr8_-_77912431 | 1.82 |
ENST00000357039.4
ENST00000522527.1 |
PEX2
|
peroxisomal biogenesis factor 2 |
| chr6_+_142623063 | 1.78 |
ENST00000296932.8
ENST00000367609.3 |
GPR126
|
G protein-coupled receptor 126 |
| chr1_+_61547405 | 1.78 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr11_+_59522837 | 1.77 |
ENST00000437946.2
|
STX3
|
syntaxin 3 |
| chr2_-_99224915 | 1.75 |
ENST00000328709.3
ENST00000409997.1 |
COA5
|
cytochrome c oxidase assembly factor 5 |
| chr20_-_45984401 | 1.74 |
ENST00000311275.7
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr17_+_40912764 | 1.74 |
ENST00000589683.1
ENST00000588928.1 |
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr5_-_88179302 | 1.66 |
ENST00000504921.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr12_+_9067327 | 1.64 |
ENST00000433083.2
ENST00000544916.1 ENST00000544539.1 ENST00000539063.1 |
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr16_-_2205352 | 1.61 |
ENST00000563192.1
|
RP11-304L19.5
|
RP11-304L19.5 |
| chr8_+_29953163 | 1.60 |
ENST00000518192.1
|
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chr2_+_109237717 | 1.59 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr17_-_76123101 | 1.59 |
ENST00000392467.3
|
TMC6
|
transmembrane channel-like 6 |
| chr16_+_2205755 | 1.58 |
ENST00000326181.6
|
TRAF7
|
TNF receptor-associated factor 7, E3 ubiquitin protein ligase |
| chr1_+_16083154 | 1.53 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr14_+_57735614 | 1.51 |
ENST00000261558.3
|
AP5M1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr21_+_17961006 | 1.51 |
ENST00000602323.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr3_-_47324242 | 1.49 |
ENST00000456548.1
ENST00000432493.1 ENST00000335044.2 ENST00000444589.2 |
KIF9
|
kinesin family member 9 |
| chr19_+_32896646 | 1.49 |
ENST00000392250.2
|
DPY19L3
|
dpy-19-like 3 (C. elegans) |
| chr3_-_149051194 | 1.47 |
ENST00000470080.1
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr14_-_23285069 | 1.45 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr6_-_33267101 | 1.44 |
ENST00000497454.1
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr19_+_32896697 | 1.44 |
ENST00000586987.1
|
DPY19L3
|
dpy-19-like 3 (C. elegans) |
| chr12_-_6772303 | 1.42 |
ENST00000396807.4
ENST00000446105.2 ENST00000341550.4 |
ING4
|
inhibitor of growth family, member 4 |
| chr12_+_96588279 | 1.41 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr2_-_75788038 | 1.40 |
ENST00000393913.3
ENST00000410113.1 |
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr2_+_33661382 | 1.40 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr3_-_47324079 | 1.38 |
ENST00000352910.4
|
KIF9
|
kinesin family member 9 |
| chr20_-_45985172 | 1.37 |
ENST00000536340.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr14_-_23285011 | 1.36 |
ENST00000397532.3
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr5_+_153570319 | 1.35 |
ENST00000377661.2
|
GALNT10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr3_-_47324008 | 1.34 |
ENST00000425853.1
|
KIF9
|
kinesin family member 9 |
| chr11_-_71791518 | 1.34 |
ENST00000537217.1
ENST00000366394.3 ENST00000358965.6 ENST00000546131.1 ENST00000543937.1 ENST00000368959.5 ENST00000541641.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr5_+_153570285 | 1.34 |
ENST00000425427.2
ENST00000297107.6 |
GALNT10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr22_-_37880543 | 1.33 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr17_+_62503147 | 1.32 |
ENST00000553412.1
|
CEP95
|
centrosomal protein 95kDa |
| chr11_-_71791435 | 1.32 |
ENST00000351960.6
ENST00000541719.1 ENST00000535111.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chrX_+_55478538 | 1.31 |
ENST00000342972.1
|
MAGEH1
|
melanoma antigen family H, 1 |
| chrX_-_119695279 | 1.30 |
ENST00000336592.6
|
CUL4B
|
cullin 4B |
| chr17_+_37844331 | 1.30 |
ENST00000578199.1
ENST00000406381.2 |
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr16_+_77225071 | 1.29 |
ENST00000439557.2
ENST00000545553.1 |
MON1B
|
MON1 secretory trafficking family member B |
| chr6_+_132455118 | 1.28 |
ENST00000458028.1
|
LINC01013
|
long intergenic non-protein coding RNA 1013 |
| chr5_+_180650271 | 1.28 |
ENST00000351937.5
ENST00000315073.5 |
TRIM41
|
tripartite motif containing 41 |
| chr3_-_128879875 | 1.27 |
ENST00000418265.1
ENST00000393292.3 ENST00000273541.8 |
ISY1-RAB43
ISY1
|
ISY1-RAB43 readthrough ISY1 splicing factor homolog (S. cerevisiae) |
| chr14_+_100531615 | 1.27 |
ENST00000392920.3
|
EVL
|
Enah/Vasp-like |
| chr1_+_156698234 | 1.26 |
ENST00000368218.4
ENST00000368216.4 |
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr17_+_47865917 | 1.24 |
ENST00000259021.4
ENST00000454930.2 ENST00000509773.1 ENST00000510819.1 ENST00000424009.2 |
KAT7
|
K(lysine) acetyltransferase 7 |
| chr4_-_89152474 | 1.24 |
ENST00000515655.1
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr14_+_63671105 | 1.23 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr12_-_105630016 | 1.23 |
ENST00000258530.3
|
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr19_+_34745442 | 1.23 |
ENST00000299505.6
ENST00000588470.1 ENST00000589583.1 ENST00000588338.2 |
KIAA0355
|
KIAA0355 |
| chr6_-_28303901 | 1.22 |
ENST00000439158.1
ENST00000446474.1 ENST00000414431.1 ENST00000344279.6 ENST00000453745.1 |
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr9_+_120466610 | 1.22 |
ENST00000394487.4
|
TLR4
|
toll-like receptor 4 |
| chr10_+_112257596 | 1.22 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5 |
| chr11_+_59522532 | 1.20 |
ENST00000337979.4
ENST00000535361.1 |
STX3
|
syntaxin 3 |
| chr5_-_88178964 | 1.17 |
ENST00000513252.1
ENST00000508569.1 ENST00000510942.1 ENST00000506554.1 |
MEF2C
|
myocyte enhancer factor 2C |
| chr10_+_35484793 | 1.16 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr2_+_148778570 | 1.16 |
ENST00000407073.1
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr2_+_170550944 | 1.16 |
ENST00000359744.3
ENST00000438838.1 ENST00000438710.1 ENST00000449906.1 ENST00000498202.2 ENST00000272797.4 |
PHOSPHO2
KLHL23
|
phosphatase, orphan 2 kelch-like family member 23 |
| chr14_+_24702073 | 1.15 |
ENST00000399440.2
|
GMPR2
|
guanosine monophosphate reductase 2 |
| chr1_+_43766642 | 1.15 |
ENST00000372476.3
|
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr21_+_18885318 | 1.14 |
ENST00000400166.1
|
CXADR
|
coxsackie virus and adenovirus receptor |
| chr5_-_88179017 | 1.14 |
ENST00000514028.1
ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr2_+_87754989 | 1.13 |
ENST00000409898.2
ENST00000419680.2 ENST00000414584.1 ENST00000455131.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr20_-_45985464 | 1.12 |
ENST00000458360.2
ENST00000262975.4 |
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr12_-_6233828 | 1.12 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr2_+_87754887 | 1.11 |
ENST00000409054.1
ENST00000331944.6 ENST00000409139.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr20_-_45985414 | 1.10 |
ENST00000461685.1
ENST00000372023.3 ENST00000540497.1 ENST00000435836.1 ENST00000471951.2 ENST00000352431.2 ENST00000396281.4 ENST00000355972.4 ENST00000360911.3 |
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr12_-_105629852 | 1.09 |
ENST00000551662.1
ENST00000553097.1 |
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr2_+_204193101 | 1.09 |
ENST00000430418.1
ENST00000424558.1 ENST00000261016.6 |
ABI2
|
abl-interactor 2 |
| chr2_+_87755054 | 1.09 |
ENST00000423846.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr4_-_4249924 | 1.08 |
ENST00000254742.2
ENST00000382753.4 ENST00000540397.1 ENST00000538516.1 |
TMEM128
|
transmembrane protein 128 |
| chr9_+_115513003 | 1.08 |
ENST00000374232.3
|
SNX30
|
sorting nexin family member 30 |
| chr14_+_24701628 | 1.08 |
ENST00000355299.4
ENST00000559836.1 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr21_+_18885430 | 1.08 |
ENST00000356275.6
ENST00000400165.1 ENST00000400169.1 ENST00000306618.10 |
CXADR
|
coxsackie virus and adenovirus receptor |
| chr6_+_34857019 | 1.08 |
ENST00000360359.3
ENST00000535627.1 |
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A |
| chr14_+_24701819 | 1.08 |
ENST00000560139.1
ENST00000559910.1 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr12_+_96588143 | 1.07 |
ENST00000228741.3
ENST00000547249.1 |
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr2_+_204193129 | 1.07 |
ENST00000417864.1
|
ABI2
|
abl-interactor 2 |
| chr11_-_71791726 | 1.07 |
ENST00000393695.3
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr1_-_17304771 | 1.06 |
ENST00000375534.3
|
MFAP2
|
microfibrillar-associated protein 2 |
| chr18_-_52989217 | 1.06 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr10_+_18948311 | 1.05 |
ENST00000377275.3
|
ARL5B
|
ADP-ribosylation factor-like 5B |
| chr4_+_2813946 | 1.04 |
ENST00000442312.2
|
SH3BP2
|
SH3-domain binding protein 2 |
| chr3_-_105588231 | 1.03 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr10_-_14996017 | 1.03 |
ENST00000378241.1
ENST00000456122.1 ENST00000418843.1 ENST00000378249.1 ENST00000396817.2 ENST00000378255.1 ENST00000378254.1 ENST00000378278.2 ENST00000357717.2 |
DCLRE1C
|
DNA cross-link repair 1C |
| chr14_+_24702127 | 1.03 |
ENST00000557854.1
ENST00000348719.7 ENST00000559104.1 ENST00000456667.3 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr14_+_75469606 | 1.03 |
ENST00000266126.5
|
EIF2B2
|
eukaryotic translation initiation factor 2B, subunit 2 beta, 39kDa |
| chr22_-_29784519 | 1.03 |
ENST00000357586.2
ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr16_-_2097787 | 1.02 |
ENST00000566380.1
ENST00000219066.1 |
NTHL1
|
nth endonuclease III-like 1 (E. coli) |
| chr15_+_96875657 | 1.02 |
ENST00000559679.1
ENST00000394171.2 |
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr14_-_90085458 | 1.02 |
ENST00000345097.4
ENST00000555855.1 ENST00000555353.1 |
FOXN3
|
forkhead box N3 |
| chr5_-_10249990 | 1.02 |
ENST00000511437.1
ENST00000280330.8 ENST00000510047.1 |
FAM173B
|
family with sequence similarity 173, member B |
| chr7_+_86781847 | 1.01 |
ENST00000432366.2
ENST00000423590.2 ENST00000394703.5 |
DMTF1
|
cyclin D binding myb-like transcription factor 1 |
| chr9_+_34652164 | 1.01 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr21_-_33984865 | 1.01 |
ENST00000458138.1
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr2_+_121010370 | 1.01 |
ENST00000420510.1
|
RALB
|
v-ral simian leukemia viral oncogene homolog B |
| chr8_+_74903580 | 1.00 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr17_-_71088797 | 0.99 |
ENST00000580557.1
ENST00000579732.1 ENST00000578620.1 ENST00000542342.2 ENST00000255559.3 ENST00000579018.1 |
SLC39A11
|
solute carrier family 39, member 11 |
| chr7_-_27219849 | 0.99 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr12_+_7055631 | 0.98 |
ENST00000543115.1
ENST00000399448.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr10_-_50747064 | 0.98 |
ENST00000355832.5
ENST00000603152.1 ENST00000447839.2 |
ERCC6
PGBD3
ERCC6-PGBD3
|
excision repair cross-complementing rodent repair deficiency, complementation group 6 piggyBac transposable element derived 3 ERCC6-PGBD3 readthrough |
| chr21_-_33984888 | 0.98 |
ENST00000382549.4
ENST00000540881.1 |
C21orf59
|
chromosome 21 open reading frame 59 |
| chr2_+_204193149 | 0.97 |
ENST00000422511.2
|
ABI2
|
abl-interactor 2 |
| chr5_+_36152163 | 0.97 |
ENST00000274255.6
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr8_-_99306564 | 0.97 |
ENST00000430223.2
|
NIPAL2
|
NIPA-like domain containing 2 |
| chr13_+_80055581 | 0.97 |
ENST00000487865.1
|
NDFIP2
|
Nedd4 family interacting protein 2 |
| chr5_+_36152091 | 0.96 |
ENST00000274254.5
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr14_+_53019993 | 0.95 |
ENST00000542169.2
ENST00000555622.1 |
GPR137C
|
G protein-coupled receptor 137C |
| chr9_+_120466650 | 0.94 |
ENST00000355622.6
|
TLR4
|
toll-like receptor 4 |
| chr14_+_56585048 | 0.94 |
ENST00000267460.4
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr15_+_77287426 | 0.94 |
ENST00000558012.1
ENST00000267939.5 ENST00000379595.3 |
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chr15_+_57511609 | 0.94 |
ENST00000543579.1
ENST00000537840.1 ENST00000343827.3 |
TCF12
|
transcription factor 12 |
| chr2_+_173686303 | 0.93 |
ENST00000397087.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr14_-_94421923 | 0.93 |
ENST00000555507.1
|
ASB2
|
ankyrin repeat and SOCS box containing 2 |
| chr14_+_53019822 | 0.93 |
ENST00000321662.6
|
GPR137C
|
G protein-coupled receptor 137C |
| chr20_+_10199468 | 0.93 |
ENST00000254976.2
ENST00000304886.2 |
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr19_-_36505098 | 0.93 |
ENST00000252984.7
ENST00000486389.1 ENST00000378875.3 ENST00000485128.1 |
ALKBH6
|
alkB, alkylation repair homolog 6 (E. coli) |
| chr1_+_22351977 | 0.93 |
ENST00000420503.1
ENST00000416769.1 ENST00000404210.2 |
LINC00339
|
long intergenic non-protein coding RNA 339 |
| chr8_-_99306611 | 0.93 |
ENST00000341166.3
|
NIPAL2
|
NIPA-like domain containing 2 |
| chr2_+_138721850 | 0.92 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chrX_-_119694538 | 0.92 |
ENST00000371322.5
|
CUL4B
|
cullin 4B |
| chr6_-_70506963 | 0.92 |
ENST00000370577.3
|
LMBRD1
|
LMBR1 domain containing 1 |
| chr14_+_70233810 | 0.91 |
ENST00000394366.2
ENST00000553548.1 ENST00000553369.1 ENST00000557154.1 ENST00000451983.2 ENST00000553635.1 |
SRSF5
|
serine/arginine-rich splicing factor 5 |
| chr20_-_57582296 | 0.91 |
ENST00000217131.5
|
CTSZ
|
cathepsin Z |
| chr5_+_138677515 | 0.91 |
ENST00000265192.4
ENST00000511706.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr10_-_18948156 | 0.91 |
ENST00000414939.1
ENST00000449529.1 ENST00000456217.1 ENST00000444660.1 |
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr10_-_14996070 | 0.91 |
ENST00000378258.1
ENST00000453695.2 ENST00000378246.2 |
DCLRE1C
|
DNA cross-link repair 1C |
| chr14_+_24702099 | 0.91 |
ENST00000420554.2
|
GMPR2
|
guanosine monophosphate reductase 2 |
| chrX_-_50557014 | 0.90 |
ENST00000376020.2
|
SHROOM4
|
shroom family member 4 |
| chr3_-_28390581 | 0.90 |
ENST00000479665.1
|
AZI2
|
5-azacytidine induced 2 |
| chr17_+_4843679 | 0.90 |
ENST00000576229.1
|
RNF167
|
ring finger protein 167 |
| chrX_+_86772787 | 0.90 |
ENST00000373114.4
|
KLHL4
|
kelch-like family member 4 |
| chr8_-_27630102 | 0.90 |
ENST00000356537.4
ENST00000522915.1 ENST00000539095.1 |
CCDC25
|
coiled-coil domain containing 25 |
| chr2_+_233925064 | 0.89 |
ENST00000359570.5
ENST00000538935.1 |
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr11_-_57282349 | 0.88 |
ENST00000528450.1
|
SLC43A1
|
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr17_-_62084241 | 0.88 |
ENST00000449662.2
|
ICAM2
|
intercellular adhesion molecule 2 |
| chr3_-_65583561 | 0.88 |
ENST00000460329.2
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr12_+_9102632 | 0.88 |
ENST00000539240.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr2_-_106013400 | 0.87 |
ENST00000409807.1
|
FHL2
|
four and a half LIM domains 2 |
| chr20_-_23066953 | 0.87 |
ENST00000246006.4
|
CD93
|
CD93 molecule |
| chr10_+_129845823 | 0.87 |
ENST00000306042.5
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chr2_-_45838374 | 0.87 |
ENST00000263736.4
|
SRBD1
|
S1 RNA binding domain 1 |
| chr9_+_139557360 | 0.87 |
ENST00000308874.7
ENST00000406555.3 ENST00000492862.2 |
EGFL7
|
EGF-like-domain, multiple 7 |
| chr17_-_76124812 | 0.86 |
ENST00000592063.1
ENST00000589271.1 ENST00000322933.4 ENST00000589553.1 |
TMC6
|
transmembrane channel-like 6 |
| chr12_-_51566562 | 0.86 |
ENST00000548108.1
|
TFCP2
|
transcription factor CP2 |
| chr11_+_134094508 | 0.86 |
ENST00000281187.5
ENST00000525095.2 |
VPS26B
|
vacuolar protein sorting 26 homolog B (S. pombe) |
| chr2_-_101767715 | 0.86 |
ENST00000376840.4
ENST00000409318.1 |
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain) |
| chr17_-_76128488 | 0.86 |
ENST00000322914.3
|
TMC6
|
transmembrane channel-like 6 |
| chr10_+_89264625 | 0.86 |
ENST00000371996.4
ENST00000371994.4 |
MINPP1
|
multiple inositol-polyphosphate phosphatase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ETS1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:1902365 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 1.1 | 3.3 | GO:0001300 | chronological cell aging(GO:0001300) |
| 1.0 | 4.0 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.9 | 6.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.9 | 2.7 | GO:0048391 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.9 | 5.3 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.8 | 5.8 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.7 | 2.2 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.7 | 2.7 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.6 | 1.9 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.6 | 3.6 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.6 | 1.8 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.6 | 1.7 | GO:0060178 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.5 | 1.6 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.5 | 4.1 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.5 | 1.9 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.5 | 1.4 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.5 | 2.3 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.5 | 0.9 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.4 | 1.3 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.4 | 13.4 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.4 | 3.7 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.4 | 1.2 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.4 | 4.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.4 | 1.4 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.3 | 2.7 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.3 | 1.7 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.3 | 1.3 | GO:1904978 | regulation of endosome organization(GO:1904978) |
| 0.3 | 0.6 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.3 | 0.9 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.3 | 3.5 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.3 | 1.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.3 | 0.8 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.3 | 0.8 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.3 | 2.2 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.3 | 1.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.3 | 1.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.3 | 6.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.3 | 1.3 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.3 | 1.0 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.3 | 1.3 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.3 | 0.8 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.3 | 1.5 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.2 | 3.0 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.2 | 0.7 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.2 | 0.5 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.2 | 0.5 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.2 | 2.7 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 0.8 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.2 | 0.6 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.2 | 1.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.2 | 0.8 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.2 | 5.7 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.2 | 1.6 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 0.2 | GO:0045399 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) |
| 0.2 | 1.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.2 | 3.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 0.8 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.2 | 0.8 | GO:1903347 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 | 0.2 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.2 | 1.2 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 | 1.7 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 0.7 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.2 | 2.2 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.2 | 1.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.2 | 0.9 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.2 | 0.9 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.2 | 0.5 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.2 | 0.2 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.2 | 0.5 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 2.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.2 | 2.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 0.6 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.2 | 0.8 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.2 | 0.5 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.2 | 0.8 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.2 | 0.8 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.2 | 1.7 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.2 | 0.5 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.1 | 0.4 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.1 | 0.7 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.1 | 0.4 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 0.4 | GO:0046832 | mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 | 0.5 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.1 | 5.2 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 0.3 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 2.9 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.1 | 1.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.1 | 0.7 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.1 | 0.5 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.1 | 0.8 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.5 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 | 0.1 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 0.9 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.7 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.1 | 0.4 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.7 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 0.3 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 0.6 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.1 | 0.9 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.3 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 | 1.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.5 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.1 | 0.3 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 1.3 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.6 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 1.2 | GO:0015886 | heme transport(GO:0015886) xenobiotic transport(GO:0042908) |
| 0.1 | 3.4 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.9 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.5 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.1 | 0.3 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 1.0 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 4.4 | GO:0048265 | response to pain(GO:0048265) |
| 0.1 | 0.6 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.1 | 0.4 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 0.6 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 1.4 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.5 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 1.6 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 0.3 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 1.2 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.8 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.7 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 1.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.8 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.7 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.1 | 0.3 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.6 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.3 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.1 | 0.7 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 0.2 | GO:0098907 | protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.1 | 1.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 2.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.3 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.2 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.2 | GO:0032679 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.1 | 0.3 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 1.0 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.7 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.3 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 | 0.7 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.1 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 1.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.4 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.6 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.2 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.1 | 0.4 | GO:0035711 | T-helper 1 cell activation(GO:0035711) |
| 0.1 | 0.6 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.3 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.1 | 0.4 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 0.8 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 0.2 | GO:0050894 | determination of affect(GO:0050894) |
| 0.1 | 0.2 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.1 | 0.3 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 0.1 | GO:0090034 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.1 | 2.5 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 | 1.1 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.1 | 0.2 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.3 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.5 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 0.5 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.1 | 0.4 | GO:0072662 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 1.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.3 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 0.8 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.5 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.1 | 0.2 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.1 | 0.5 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.1 | 1.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.7 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 1.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.2 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.1 | 0.3 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 0.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.2 | GO:0052056 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.1 | 0.5 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.1 | 1.0 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 0.2 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.1 | 0.2 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.1 | 0.2 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.1 | 0.5 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.5 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.1 | 0.1 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.1 | 0.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.3 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.1 | 0.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.2 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.0 | 0.6 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 1.2 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.6 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.2 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.5 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 1.7 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.6 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.3 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.9 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.4 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.6 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.3 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.8 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 1.3 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.4 | GO:0090043 | tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.3 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.5 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.5 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 1.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.5 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 2.3 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.2 | GO:0071484 | cellular response to light intensity(GO:0071484) |
| 0.0 | 0.9 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.4 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.7 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.4 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.4 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.2 | GO:0060266 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 1.0 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.1 | GO:2000547 | myeloid dendritic cell chemotaxis(GO:0002408) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 0.8 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.2 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.7 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.2 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.3 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.1 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.8 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 1.3 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.3 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 1.2 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 1.2 | GO:0042269 | regulation of natural killer cell mediated cytotoxicity(GO:0042269) |
| 0.0 | 0.5 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.6 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 1.0 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.2 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.2 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.0 | GO:1902616 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.8 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.5 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.0 | 0.4 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.2 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.5 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.4 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0097490 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.0 | 0.9 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.2 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.7 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.0 | 0.2 | GO:2000010 | positive regulation of protein localization to cell surface(GO:2000010) |
| 0.0 | 0.5 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.2 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 2.1 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 0.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 1.7 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.3 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.1 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.3 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.1 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.5 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.3 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.4 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.4 | GO:0046051 | UTP metabolic process(GO:0046051) |
| 0.0 | 0.5 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.3 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 1.4 | GO:1901880 | negative regulation of protein depolymerization(GO:1901880) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.4 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.8 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.9 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 1.5 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.0 | 1.2 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.2 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.3 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.2 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 0.0 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.7 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.1 | GO:0072079 | nephron tubule formation(GO:0072079) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 1.0 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.9 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.1 | GO:0072319 | synaptic vesicle uncoating(GO:0016191) vesicle uncoating(GO:0072319) |
| 0.0 | 0.2 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.3 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.3 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.3 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.0 | 1.3 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.0 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.0 | 0.2 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 | 1.4 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.2 | GO:0097502 | mannosylation(GO:0097502) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.8 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.2 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.2 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.5 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.6 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.5 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.7 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.7 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.4 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 0.7 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.0 | 1.3 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.4 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.4 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.0 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.3 | GO:0003091 | renal water homeostasis(GO:0003091) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.4 | GO:0006266 | DNA ligation(GO:0006266) |
| 0.0 | 1.4 | GO:0042633 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.0 | 0.1 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.0 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.4 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.0 | GO:0097354 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:1900225 | regulation of NLRP3 inflammasome complex assembly(GO:1900225) positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0055028 | cortical microtubule(GO:0055028) |
| 1.2 | 3.7 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 1.2 | 4.8 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.9 | 5.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.7 | 3.6 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.6 | 3.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.5 | 3.7 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.3 | 7.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 1.3 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.3 | 1.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.3 | 2.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.2 | 0.7 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 0.7 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.2 | 0.9 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.2 | 0.6 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.2 | 3.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 2.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 2.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 0.9 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.2 | 0.5 | GO:0042022 | interleukin-12 receptor complex(GO:0042022) |
| 0.2 | 0.5 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.2 | 3.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 2.5 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.2 | 0.9 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 5.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 5.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.0 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 3.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.9 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.4 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.4 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 1.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.5 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.8 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 2.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.3 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 0.4 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.1 | 0.5 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 3.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.8 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.5 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 0.3 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 1.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.6 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 1.8 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 0.8 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.8 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.6 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 1.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.6 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 5.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.2 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.1 | 4.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 0.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 1.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.6 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 1.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.8 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.6 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 2.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 2.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 0.2 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.1 | 0.2 | GO:0032449 | CBM complex(GO:0032449) |
| 0.1 | 4.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 1.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 0.3 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 1.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 0.7 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.4 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.1 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.0 | 0.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 1.0 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 4.0 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 2.0 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.4 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.4 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 1.1 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.5 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 1.4 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 1.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 2.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.0 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.5 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 1.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.8 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 1.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.9 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 2.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 3.1 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 2.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 8.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.9 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 1.3 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 6.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 6.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.2 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.0 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.3 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 2.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.2 | GO:0097342 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 1.1 | GO:0005776 | autophagosome(GO:0005776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.0 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 1.1 | 11.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.9 | 5.2 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.8 | 2.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.6 | 1.9 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.5 | 3.7 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.5 | 3.7 | GO:0005534 | galactose binding(GO:0005534) |
| 0.4 | 1.3 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.4 | 4.6 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.4 | 6.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 3.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.3 | 1.0 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.3 | 2.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.3 | 1.3 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.3 | 1.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.3 | 1.6 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.3 | 0.9 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.3 | 1.2 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.3 | 8.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.3 | 0.9 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.3 | 1.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.3 | 0.8 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.3 | 4.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.3 | 0.8 | GO:0019961 | interferon binding(GO:0019961) |
| 0.3 | 0.8 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.3 | 0.8 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.3 | 0.8 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.3 | 0.8 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.2 | 3.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 1.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 1.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 2.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 0.7 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.2 | 5.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 2.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 2.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 1.0 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.2 | 2.8 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.2 | 0.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.2 | 0.7 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.2 | 0.5 | GO:0016517 | interleukin-12 receptor activity(GO:0016517) |
| 0.2 | 0.7 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.2 | 0.5 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.2 | 0.5 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.2 | 1.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 0.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 0.8 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.2 | 1.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.2 | 4.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 0.7 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.9 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.7 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 1.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 1.0 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 1.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 2.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 4.0 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 1.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 1.5 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) |
| 0.1 | 1.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.5 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.4 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 0.3 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.1 | 6.0 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 2.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.9 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.1 | 0.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 2.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.4 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.4 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 0.3 | GO:0001003 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.1 | 2.5 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 1.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.9 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.3 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 0.3 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.5 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.4 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.6 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.1 | 0.8 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.6 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.4 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 0.7 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.2 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.1 | 1.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 1.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 2.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 1.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.9 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.2 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.1 | 0.3 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 1.4 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 1.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 0.7 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 6.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.2 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.1 | 1.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.3 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.2 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 0.8 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.9 | GO:0005346 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.1 | 0.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.2 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 1.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 1.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 1.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.3 | GO:0001016 | RNA polymerase III regulatory region DNA binding(GO:0001016) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.3 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.3 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 2.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.3 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 1.1 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 1.1 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.3 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.0 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.6 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.7 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.0 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.4 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.3 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.4 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0004663 | protein geranylgeranyltransferase activity(GO:0004661) Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 1.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.9 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.5 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.7 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 1.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.4 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.9 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.5 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.6 | GO:0015149 | hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.4 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.5 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.0 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 1.1 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 1.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.6 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 1.4 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.0 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 1.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 3.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 0.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 9.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 0.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 3.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 3.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 0.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 4.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 0.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 3.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 2.9 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 2.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 3.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 2.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.0 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 1.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 3.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 7.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 1.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 2.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.1 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 2.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 5.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 2.8 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 6.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 3.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 3.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 4.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 2.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 3.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 4.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.1 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 4.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.0 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 1.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 3.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 2.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.8 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 1.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 1.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 0.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 0.8 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 3.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 0.8 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.1 | 1.7 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.4 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 2.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 2.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 3.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.5 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.0 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 1.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 2.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.0 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |