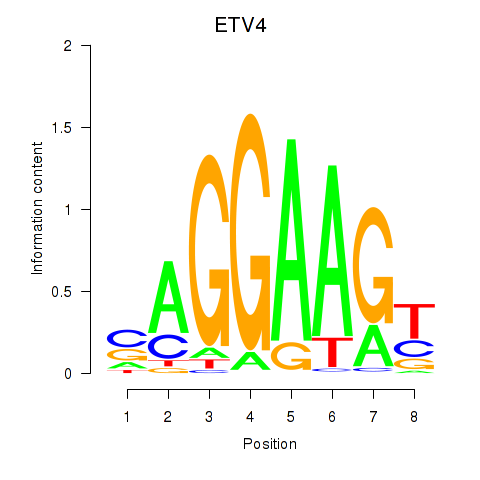
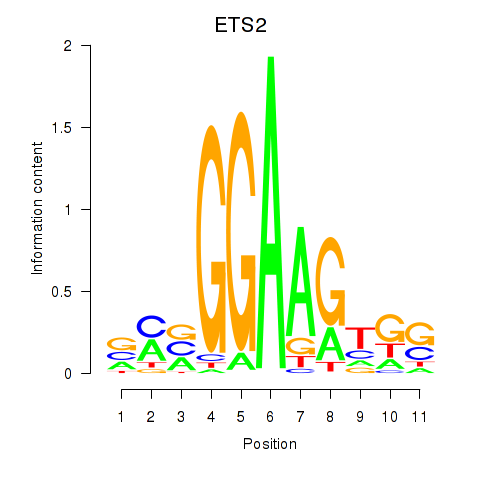
Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for ETV4_ETS2
Z-value: 0.73
Transcription factors associated with ETV4_ETS2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV4
|
ENSG00000175832.8 | ETS variant transcription factor 4 |
|
ETS2
|
ENSG00000157557.7 | ETS proto-oncogene 2, transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ETS2 | hg19_v2_chr21_+_40177143_40177231, hg19_v2_chr21_+_40177755_40177875 | 0.41 | 4.3e-02 | Click! |
| ETV4 | hg19_v2_chr17_-_41623716_41623800, hg19_v2_chr17_-_41623009_41623053 | 0.38 | 5.8e-02 | Click! |
Activity profile of ETV4_ETS2 motif
Sorted Z-values of ETV4_ETS2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_71184931 | 1.58 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr15_+_71185148 | 1.51 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr5_+_125758813 | 1.43 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr5_+_125758865 | 1.41 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr1_-_173174681 | 1.41 |
ENST00000367718.1
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr21_+_26934165 | 1.40 |
ENST00000456917.1
|
MIR155HG
|
MIR155 host gene (non-protein coding) |
| chr2_+_103035102 | 1.39 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr5_-_131826457 | 1.32 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chr7_+_108210012 | 1.27 |
ENST00000249356.3
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr9_+_35732312 | 1.14 |
ENST00000353704.2
|
CREB3
|
cAMP responsive element binding protein 3 |
| chr8_-_104427313 | 1.09 |
ENST00000297578.4
|
SLC25A32
|
solute carrier family 25 (mitochondrial folate carrier), member 32 |
| chr20_+_44746939 | 1.08 |
ENST00000372276.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr2_-_225811747 | 1.02 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr6_+_126240442 | 1.02 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr16_-_67969888 | 1.02 |
ENST00000574576.2
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr12_+_100660909 | 1.00 |
ENST00000549687.1
|
SCYL2
|
SCY1-like 2 (S. cerevisiae) |
| chr19_+_10197463 | 1.00 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr11_-_3862059 | 0.99 |
ENST00000396978.1
|
RHOG
|
ras homolog family member G |
| chr3_+_47324424 | 0.99 |
ENST00000437353.1
ENST00000232766.5 ENST00000455924.2 |
KLHL18
|
kelch-like family member 18 |
| chr15_-_80263506 | 0.95 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr15_+_75074410 | 0.94 |
ENST00000439220.2
|
CSK
|
c-src tyrosine kinase |
| chr11_+_2421718 | 0.94 |
ENST00000380996.5
ENST00000333256.6 ENST00000380992.1 ENST00000437110.1 ENST00000435795.1 |
TSSC4
|
tumor suppressing subtransferable candidate 4 |
| chr20_+_44746885 | 0.94 |
ENST00000372285.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr2_+_32853093 | 0.89 |
ENST00000448773.1
ENST00000317907.4 |
TTC27
|
tetratricopeptide repeat domain 27 |
| chr10_-_49812997 | 0.88 |
ENST00000417912.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr6_+_31553901 | 0.84 |
ENST00000418507.2
ENST00000438075.2 ENST00000376100.3 ENST00000376111.4 |
LST1
|
leukocyte specific transcript 1 |
| chr6_+_31553978 | 0.84 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr6_+_43603552 | 0.83 |
ENST00000372171.4
|
MAD2L1BP
|
MAD2L1 binding protein |
| chr1_-_150738261 | 0.81 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr15_+_75074385 | 0.80 |
ENST00000220003.9
|
CSK
|
c-src tyrosine kinase |
| chr16_+_67261008 | 0.79 |
ENST00000304800.9
ENST00000563953.1 ENST00000565201.1 |
TMEM208
|
transmembrane protein 208 |
| chr21_+_43619796 | 0.78 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr6_+_31515337 | 0.77 |
ENST00000376148.4
ENST00000376145.4 |
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr16_+_57673430 | 0.77 |
ENST00000540164.2
ENST00000568531.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr17_+_26662597 | 0.76 |
ENST00000544907.2
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr11_-_46722117 | 0.75 |
ENST00000311956.4
|
ARHGAP1
|
Rho GTPase activating protein 1 |
| chr2_+_228678550 | 0.74 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr16_+_2732476 | 0.74 |
ENST00000301738.4
ENST00000564195.1 |
KCTD5
|
potassium channel tetramerization domain containing 5 |
| chr14_-_45431091 | 0.73 |
ENST00000579157.1
ENST00000396128.4 ENST00000556500.1 |
KLHL28
|
kelch-like family member 28 |
| chr6_-_33290580 | 0.73 |
ENST00000446511.1
ENST00000446403.1 ENST00000414083.2 ENST00000266000.6 ENST00000374542.5 |
DAXX
|
death-domain associated protein |
| chr3_-_112693865 | 0.71 |
ENST00000471858.1
ENST00000295863.4 ENST00000308611.3 |
CD200R1
|
CD200 receptor 1 |
| chr1_-_101360331 | 0.71 |
ENST00000416479.1
ENST00000370113.3 |
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr12_+_108908962 | 0.71 |
ENST00000552695.1
ENST00000552758.1 ENST00000361549.2 |
FICD
|
FIC domain containing |
| chr10_-_49813090 | 0.70 |
ENST00000249601.4
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr19_-_4338783 | 0.70 |
ENST00000601482.1
ENST00000600324.1 |
STAP2
|
signal transducing adaptor family member 2 |
| chr11_-_72432950 | 0.70 |
ENST00000426523.1
ENST00000429686.1 |
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr14_+_77924204 | 0.69 |
ENST00000555133.1
|
AHSA1
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast) |
| chr14_+_77924373 | 0.68 |
ENST00000216479.3
ENST00000535854.2 ENST00000555517.1 |
AHSA1
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast) |
| chr17_+_26662730 | 0.68 |
ENST00000226225.2
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr17_+_32582293 | 0.67 |
ENST00000580907.1
ENST00000225831.4 |
CCL2
|
chemokine (C-C motif) ligand 2 |
| chr3_-_172428959 | 0.67 |
ENST00000475381.1
ENST00000538775.1 ENST00000273512.3 ENST00000543711.1 |
NCEH1
|
neutral cholesterol ester hydrolase 1 |
| chr16_+_57653989 | 0.64 |
ENST00000567835.1
ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr14_+_73603126 | 0.64 |
ENST00000557356.1
ENST00000556864.1 ENST00000556533.1 ENST00000556951.1 ENST00000557293.1 ENST00000553719.1 ENST00000553599.1 ENST00000556011.1 ENST00000394157.3 ENST00000357710.4 ENST00000324501.5 ENST00000560005.2 ENST00000555254.1 ENST00000261970.3 ENST00000344094.3 ENST00000554131.1 ENST00000557037.1 |
PSEN1
|
presenilin 1 |
| chr16_+_57653854 | 0.63 |
ENST00000568908.1
ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr6_-_43595039 | 0.63 |
ENST00000307114.7
|
GTPBP2
|
GTP binding protein 2 |
| chr16_+_56485402 | 0.62 |
ENST00000566157.1
ENST00000562150.1 ENST00000561646.1 ENST00000568397.1 |
OGFOD1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr6_-_11382478 | 0.61 |
ENST00000397378.3
ENST00000513989.1 ENST00000508546.1 ENST00000504387.1 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr17_-_18218270 | 0.61 |
ENST00000321105.5
|
TOP3A
|
topoisomerase (DNA) III alpha |
| chr1_-_894620 | 0.61 |
ENST00000327044.6
|
NOC2L
|
nucleolar complex associated 2 homolog (S. cerevisiae) |
| chr20_-_48532019 | 0.61 |
ENST00000289431.5
|
SPATA2
|
spermatogenesis associated 2 |
| chr11_-_3862206 | 0.59 |
ENST00000351018.4
|
RHOG
|
ras homolog family member G |
| chr17_+_7461613 | 0.59 |
ENST00000438470.1
ENST00000436057.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr5_+_140071011 | 0.59 |
ENST00000230771.3
ENST00000509299.1 ENST00000503873.1 ENST00000435019.2 ENST00000437649.2 ENST00000432671.2 |
HARS2
|
histidyl-tRNA synthetase 2, mitochondrial |
| chr17_-_56595196 | 0.58 |
ENST00000579921.1
ENST00000579925.1 ENST00000323456.5 |
MTMR4
|
myotubularin related protein 4 |
| chr19_-_4338838 | 0.58 |
ENST00000594605.1
|
STAP2
|
signal transducing adaptor family member 2 |
| chr16_+_31085714 | 0.58 |
ENST00000300850.5
ENST00000564189.1 ENST00000428260.1 |
ZNF646
|
zinc finger protein 646 |
| chr1_-_160313025 | 0.57 |
ENST00000368069.3
ENST00000241704.7 |
COPA
|
coatomer protein complex, subunit alpha |
| chr4_-_139163491 | 0.57 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr17_+_18218587 | 0.56 |
ENST00000406438.3
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8 |
| chr17_-_20370847 | 0.56 |
ENST00000423676.3
ENST00000324290.5 |
LGALS9B
|
lectin, galactoside-binding, soluble, 9B |
| chr3_-_107809816 | 0.56 |
ENST00000361309.5
ENST00000355354.7 |
CD47
|
CD47 molecule |
| chr4_+_103423055 | 0.54 |
ENST00000505458.1
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr16_+_31044812 | 0.54 |
ENST00000313843.3
|
STX4
|
syntaxin 4 |
| chr6_+_127898312 | 0.54 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr8_+_86089460 | 0.54 |
ENST00000418930.2
|
E2F5
|
E2F transcription factor 5, p130-binding |
| chr1_+_16767167 | 0.54 |
ENST00000337132.5
|
NECAP2
|
NECAP endocytosis associated 2 |
| chr2_-_153573965 | 0.53 |
ENST00000448428.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr7_-_5569588 | 0.53 |
ENST00000417101.1
|
ACTB
|
actin, beta |
| chr5_-_138862326 | 0.53 |
ENST00000330794.4
|
TMEM173
|
transmembrane protein 173 |
| chr5_-_176730676 | 0.51 |
ENST00000393611.2
ENST00000303251.6 ENST00000303270.6 |
RAB24
|
RAB24, member RAS oncogene family |
| chr4_+_110736659 | 0.51 |
ENST00000394631.3
ENST00000226796.6 |
GAR1
|
GAR1 ribonucleoprotein |
| chr19_-_56632592 | 0.49 |
ENST00000587279.1
ENST00000270459.3 |
ZNF787
|
zinc finger protein 787 |
| chr5_-_150466692 | 0.49 |
ENST00000315050.7
ENST00000523338.1 ENST00000522100.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr9_-_132404374 | 0.49 |
ENST00000277459.4
ENST00000450050.2 ENST00000277458.4 |
ASB6
|
ankyrin repeat and SOCS box containing 6 |
| chr7_+_55086794 | 0.49 |
ENST00000275493.2
ENST00000442591.1 |
EGFR
|
epidermal growth factor receptor |
| chr5_+_68530668 | 0.49 |
ENST00000506563.1
|
CDK7
|
cyclin-dependent kinase 7 |
| chr1_+_212738676 | 0.49 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr10_-_96122682 | 0.49 |
ENST00000371361.3
|
NOC3L
|
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr11_+_19799327 | 0.49 |
ENST00000540292.1
|
NAV2
|
neuron navigator 2 |
| chr5_+_176730769 | 0.48 |
ENST00000303204.4
ENST00000503216.1 |
PRELID1
|
PRELI domain containing 1 |
| chr14_-_53417732 | 0.48 |
ENST00000399304.3
ENST00000395631.2 ENST00000341590.3 ENST00000343279.4 |
FERMT2
|
fermitin family member 2 |
| chr1_-_207095324 | 0.48 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr4_-_168155417 | 0.47 |
ENST00000511269.1
ENST00000506697.1 ENST00000512042.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr9_-_136242909 | 0.47 |
ENST00000371991.3
ENST00000545297.1 |
SURF4
|
surfeit 4 |
| chr1_+_16767195 | 0.47 |
ENST00000504551.2
ENST00000457722.2 ENST00000406746.1 ENST00000443980.2 |
NECAP2
|
NECAP endocytosis associated 2 |
| chr5_+_133984462 | 0.47 |
ENST00000398844.2
ENST00000322887.4 |
SEC24A
|
SEC24 family member A |
| chr2_-_37193606 | 0.47 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chr3_-_11762202 | 0.47 |
ENST00000445411.1
ENST00000404339.1 ENST00000273038.3 |
VGLL4
|
vestigial like 4 (Drosophila) |
| chr6_+_31554636 | 0.46 |
ENST00000433492.1
|
LST1
|
leukocyte specific transcript 1 |
| chr17_+_45908974 | 0.46 |
ENST00000269025.4
|
LRRC46
|
leucine rich repeat containing 46 |
| chr16_-_67260901 | 0.46 |
ENST00000341546.3
ENST00000409509.1 ENST00000433915.1 ENST00000454102.2 |
LRRC29
AC040160.1
|
leucine rich repeat containing 29 Uncharacterized protein; cDNA FLJ57407, weakly similar to Mus musculus leucine rich repeat containing 29 (Lrrc29), mRNA |
| chr20_-_48532046 | 0.46 |
ENST00000543716.1
|
SPATA2
|
spermatogenesis associated 2 |
| chr2_+_118846008 | 0.46 |
ENST00000245787.4
|
INSIG2
|
insulin induced gene 2 |
| chr12_-_10151773 | 0.45 |
ENST00000298527.6
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1, member B |
| chr6_+_31554826 | 0.45 |
ENST00000376089.2
ENST00000396112.2 |
LST1
|
leukocyte specific transcript 1 |
| chr8_+_38585704 | 0.45 |
ENST00000519416.1
ENST00000520615.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr2_+_169659121 | 0.45 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr19_-_59070239 | 0.45 |
ENST00000595957.1
ENST00000253023.3 |
UBE2M
|
ubiquitin-conjugating enzyme E2M |
| chr17_-_74722536 | 0.45 |
ENST00000585429.1
|
JMJD6
|
jumonji domain containing 6 |
| chr20_-_2451395 | 0.44 |
ENST00000339610.6
ENST00000381342.2 ENST00000438552.2 |
SNRPB
|
small nuclear ribonucleoprotein polypeptides B and B1 |
| chr17_+_41561317 | 0.44 |
ENST00000540306.1
ENST00000262415.3 ENST00000605777.1 |
DHX8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr3_+_38080691 | 0.43 |
ENST00000308059.6
ENST00000346219.3 ENST00000452631.2 |
DLEC1
|
deleted in lung and esophageal cancer 1 |
| chr1_+_152881014 | 0.43 |
ENST00000368764.3
ENST00000392667.2 |
IVL
|
involucrin |
| chr11_+_6502675 | 0.43 |
ENST00000254616.6
ENST00000530751.1 |
TIMM10B
|
translocase of inner mitochondrial membrane 10 homolog B (yeast) |
| chr6_+_29364416 | 0.43 |
ENST00000383555.2
|
OR12D2
|
olfactory receptor, family 12, subfamily D, member 2 (gene/pseudogene) |
| chr15_-_41120896 | 0.43 |
ENST00000299174.5
ENST00000427255.2 |
PPP1R14D
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr5_+_35856951 | 0.43 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr11_-_3078838 | 0.42 |
ENST00000397111.5
|
CARS
|
cysteinyl-tRNA synthetase |
| chr16_+_84682108 | 0.42 |
ENST00000564996.1
ENST00000258157.5 ENST00000567410.1 |
KLHL36
|
kelch-like family member 36 |
| chr8_+_86089619 | 0.42 |
ENST00000256117.5
ENST00000416274.2 |
E2F5
|
E2F transcription factor 5, p130-binding |
| chr4_-_168155577 | 0.42 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr13_+_46039037 | 0.42 |
ENST00000349995.5
|
COG3
|
component of oligomeric golgi complex 3 |
| chr10_-_43892279 | 0.42 |
ENST00000443950.2
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr15_+_81589254 | 0.42 |
ENST00000394652.2
|
IL16
|
interleukin 16 |
| chr4_+_8201091 | 0.42 |
ENST00000382521.3
ENST00000245105.3 ENST00000457650.2 ENST00000539824.1 |
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr17_+_7255208 | 0.41 |
ENST00000333751.3
|
KCTD11
|
potassium channel tetramerization domain containing 11 |
| chr1_+_169337172 | 0.41 |
ENST00000367807.3
ENST00000367808.3 ENST00000329281.2 ENST00000420531.1 |
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr1_-_161102367 | 0.41 |
ENST00000464113.1
|
DEDD
|
death effector domain containing |
| chr1_-_112046289 | 0.41 |
ENST00000241356.4
|
ADORA3
|
adenosine A3 receptor |
| chr12_+_7079944 | 0.41 |
ENST00000261406.6
|
EMG1
|
EMG1 N1-specific pseudouridine methyltransferase |
| chr17_+_1674982 | 0.40 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr5_-_892648 | 0.40 |
ENST00000483173.1
ENST00000435709.2 |
BRD9
|
bromodomain containing 9 |
| chr1_-_109618566 | 0.40 |
ENST00000338366.5
|
TAF13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr5_+_68530697 | 0.40 |
ENST00000256443.3
ENST00000514676.1 |
CDK7
|
cyclin-dependent kinase 7 |
| chrX_+_153775821 | 0.39 |
ENST00000263518.6
ENST00000470142.1 ENST00000393549.2 ENST00000455588.2 ENST00000369602.3 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr8_+_104311059 | 0.39 |
ENST00000358755.4
ENST00000523739.1 ENST00000540287.1 |
FZD6
|
frizzled family receptor 6 |
| chr6_-_155635583 | 0.39 |
ENST00000367166.4
|
TFB1M
|
transcription factor B1, mitochondrial |
| chr3_-_51975942 | 0.39 |
ENST00000232888.6
|
RRP9
|
ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast) |
| chr22_+_37309662 | 0.39 |
ENST00000403662.3
ENST00000262825.5 |
CSF2RB
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr12_+_112563335 | 0.39 |
ENST00000549358.1
ENST00000257604.5 ENST00000548092.1 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr2_-_163175133 | 0.39 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr12_+_112563303 | 0.39 |
ENST00000412615.2
|
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr11_-_70507867 | 0.39 |
ENST00000412252.1
ENST00000409161.1 ENST00000409530.1 |
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr3_+_119013185 | 0.39 |
ENST00000264245.4
|
ARHGAP31
|
Rho GTPase activating protein 31 |
| chr1_-_38273840 | 0.38 |
ENST00000373044.2
|
YRDC
|
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr12_+_117348742 | 0.38 |
ENST00000309909.5
ENST00000455858.2 |
FBXW8
|
F-box and WD repeat domain containing 8 |
| chr1_-_161102421 | 0.38 |
ENST00000490843.2
ENST00000368006.3 ENST00000392188.1 ENST00000545495.1 |
DEDD
|
death effector domain containing |
| chr2_-_219134343 | 0.38 |
ENST00000447885.1
ENST00000420660.1 |
AAMP
|
angio-associated, migratory cell protein |
| chr2_+_234160340 | 0.38 |
ENST00000417017.1
ENST00000392020.4 ENST00000392018.1 |
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr2_-_153573887 | 0.37 |
ENST00000493468.2
ENST00000545856.1 |
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr4_-_109089573 | 0.37 |
ENST00000265165.1
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr10_-_43892668 | 0.37 |
ENST00000544000.1
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr17_-_74722672 | 0.37 |
ENST00000397625.4
ENST00000445478.2 |
JMJD6
|
jumonji domain containing 6 |
| chr20_+_49575342 | 0.37 |
ENST00000244051.1
|
MOCS3
|
molybdenum cofactor synthesis 3 |
| chr19_+_16308711 | 0.37 |
ENST00000429941.2
ENST00000444449.2 ENST00000589822.1 |
AP1M1
|
adaptor-related protein complex 1, mu 1 subunit |
| chr11_-_116658758 | 0.37 |
ENST00000227322.3
|
ZNF259
|
zinc finger protein 259 |
| chr6_-_31620403 | 0.37 |
ENST00000451898.1
ENST00000439687.2 ENST00000362049.6 ENST00000424480.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr5_-_114961673 | 0.36 |
ENST00000333314.3
|
TMED7-TICAM2
|
TMED7-TICAM2 readthrough |
| chr7_-_108209897 | 0.36 |
ENST00000313516.5
|
THAP5
|
THAP domain containing 5 |
| chr11_+_65479702 | 0.36 |
ENST00000530446.1
ENST00000534104.1 ENST00000530605.1 ENST00000528198.1 ENST00000531880.1 ENST00000534650.1 |
KAT5
|
K(lysine) acetyltransferase 5 |
| chr6_-_28411241 | 0.36 |
ENST00000289788.4
|
ZSCAN23
|
zinc finger and SCAN domain containing 23 |
| chr6_+_31554612 | 0.36 |
ENST00000211921.7
|
LST1
|
leukocyte specific transcript 1 |
| chr16_+_57220193 | 0.36 |
ENST00000564435.1
ENST00000562959.1 ENST00000394420.4 ENST00000568505.2 ENST00000537866.1 |
RSPRY1
|
ring finger and SPRY domain containing 1 |
| chr14_+_24630465 | 0.36 |
ENST00000557894.1
ENST00000559284.1 ENST00000560275.1 |
IRF9
|
interferon regulatory factor 9 |
| chr18_+_72265084 | 0.36 |
ENST00000582337.1
|
ZNF407
|
zinc finger protein 407 |
| chr21_+_43933946 | 0.35 |
ENST00000352133.2
|
SLC37A1
|
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
| chr1_-_118472171 | 0.35 |
ENST00000369442.3
|
GDAP2
|
ganglioside induced differentiation associated protein 2 |
| chr9_-_33473882 | 0.35 |
ENST00000455041.2
ENST00000353159.2 ENST00000297990.4 ENST00000379471.2 |
NOL6
|
nucleolar protein 6 (RNA-associated) |
| chr2_+_65454926 | 0.35 |
ENST00000542850.1
ENST00000377982.4 |
ACTR2
|
ARP2 actin-related protein 2 homolog (yeast) |
| chr17_+_5390220 | 0.35 |
ENST00000381165.3
|
MIS12
|
MIS12 kinetochore complex component |
| chr3_+_14693247 | 0.35 |
ENST00000383794.3
ENST00000303688.7 |
CCDC174
|
coiled-coil domain containing 174 |
| chrX_+_15525426 | 0.35 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr9_+_116037922 | 0.35 |
ENST00000374198.4
|
PRPF4
|
pre-mRNA processing factor 4 |
| chr3_-_71179988 | 0.35 |
ENST00000491238.1
|
FOXP1
|
forkhead box P1 |
| chr17_+_7155343 | 0.35 |
ENST00000573513.1
ENST00000354429.2 ENST00000574255.1 ENST00000396627.2 ENST00000356683.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr6_+_31554456 | 0.35 |
ENST00000339530.4
|
LST1
|
leukocyte specific transcript 1 |
| chr1_-_153513170 | 0.34 |
ENST00000368717.2
|
S100A5
|
S100 calcium binding protein A5 |
| chr1_-_111746966 | 0.34 |
ENST00000369752.5
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr12_+_132195617 | 0.34 |
ENST00000261674.4
ENST00000535236.1 ENST00000541286.1 |
SFSWAP
|
splicing factor, suppressor of white-apricot homolog (Drosophila) |
| chr2_+_105050794 | 0.34 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr11_-_116658695 | 0.34 |
ENST00000429220.1
ENST00000444935.1 |
ZNF259
|
zinc finger protein 259 |
| chr8_+_38586068 | 0.34 |
ENST00000443286.2
ENST00000520340.1 ENST00000518415.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr11_-_70507901 | 0.34 |
ENST00000449833.2
ENST00000357171.3 ENST00000449116.2 |
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr18_+_55862622 | 0.34 |
ENST00000456173.2
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr16_+_8891670 | 0.34 |
ENST00000268261.4
ENST00000539622.1 ENST00000569958.1 ENST00000537352.1 |
PMM2
|
phosphomannomutase 2 |
| chr3_-_71179699 | 0.34 |
ENST00000497355.1
|
FOXP1
|
forkhead box P1 |
| chr7_+_157129660 | 0.34 |
ENST00000429029.2
ENST00000262177.4 ENST00000417758.1 ENST00000452797.2 ENST00000443280.1 |
DNAJB6
|
DnaJ (Hsp40) homolog, subfamily B, member 6 |
| chr6_-_34664612 | 0.34 |
ENST00000374023.3
ENST00000374026.3 |
C6orf106
|
chromosome 6 open reading frame 106 |
| chr2_+_234160217 | 0.33 |
ENST00000392017.4
ENST00000347464.5 ENST00000444735.1 ENST00000373525.5 ENST00000419681.1 |
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr15_-_62457480 | 0.33 |
ENST00000380392.3
|
C2CD4B
|
C2 calcium-dependent domain containing 4B |
| chr21_-_34186006 | 0.33 |
ENST00000490358.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr2_-_24583583 | 0.33 |
ENST00000355123.4
|
ITSN2
|
intersectin 2 |
| chr12_+_121570631 | 0.33 |
ENST00000546057.1
ENST00000377162.2 ENST00000328963.5 ENST00000535250.1 ENST00000541446.1 |
P2RX7
|
purinergic receptor P2X, ligand-gated ion channel, 7 |
| chr5_+_149380256 | 0.33 |
ENST00000502717.1
|
HMGXB3
|
HMG box domain containing 3 |
| chr17_-_47045949 | 0.32 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide |
| chr8_+_104426942 | 0.32 |
ENST00000297579.5
|
DCAF13
|
DDB1 and CUL4 associated factor 13 |
| chr17_-_18218237 | 0.32 |
ENST00000542570.1
|
TOP3A
|
topoisomerase (DNA) III alpha |
| chr10_+_115439699 | 0.32 |
ENST00000369315.1
|
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr6_+_43739697 | 0.32 |
ENST00000230480.6
|
VEGFA
|
vascular endothelial growth factor A |
| chr3_-_11685345 | 0.32 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of ETV4_ETS2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.4 | 1.2 | GO:0015883 | FAD transport(GO:0015883) FAD transmembrane transport(GO:0035350) |
| 0.4 | 1.2 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.4 | 2.7 | GO:0033590 | response to cobalamin(GO:0033590) |
| 0.3 | 0.3 | GO:0043132 | NAD transport(GO:0043132) |
| 0.3 | 0.8 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.3 | 0.8 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.2 | 1.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.2 | 0.7 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.2 | 1.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.2 | 0.7 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.2 | 0.7 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.2 | 1.7 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.2 | 1.5 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.2 | 0.8 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.2 | 0.7 | GO:1903935 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.2 | 0.5 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.2 | 0.8 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.1 | 0.1 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 0.4 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.1 | 0.7 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.4 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.1 | 0.3 | GO:1902566 | regulation of eosinophil activation(GO:1902566) |
| 0.1 | 0.7 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.4 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.1 | 0.5 | GO:2000630 | positive regulation of miRNA metabolic process(GO:2000630) |
| 0.1 | 0.5 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.4 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 0.5 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.4 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.2 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.1 | 0.5 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.1 | 0.4 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.1 | 0.1 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 0.7 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.3 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.6 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.1 | 0.3 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.3 | GO:0060381 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.1 | 0.3 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.1 | 0.3 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.1 | 0.4 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.4 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 0.5 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.1 | 0.3 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.3 | GO:0042704 | detection of oxygen(GO:0003032) uterine wall breakdown(GO:0042704) |
| 0.1 | 0.6 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 0.3 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.5 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.4 | GO:0035936 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.1 | 0.5 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.1 | 0.8 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.3 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.1 | GO:0034125 | negative regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034125) |
| 0.1 | 0.3 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.1 | 0.3 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 0.3 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.1 | 1.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.2 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.1 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.2 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) positive regulation of oocyte maturation(GO:1900195) |
| 0.1 | 0.4 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.2 | GO:0061110 | dense core granule biogenesis(GO:0061110) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.6 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 0.2 | GO:0035668 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.1 | 0.4 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 | 0.3 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.3 | GO:0097325 | negative regulation of extracellular matrix disassembly(GO:0010716) melanocyte proliferation(GO:0097325) |
| 0.1 | 0.2 | GO:0050894 | determination of affect(GO:0050894) |
| 0.1 | 0.2 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.2 | GO:2000283 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) regulation of cellular amino acid biosynthetic process(GO:2000282) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.1 | 0.2 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.1 | 0.1 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.1 | 0.2 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
| 0.1 | 0.3 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) response to hypobaric hypoxia(GO:1990910) |
| 0.1 | 0.3 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.1 | 0.3 | GO:0035720 | intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 1.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.2 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.1 | 0.2 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.1 | 0.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.3 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.5 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.2 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.1 | 0.3 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.1 | 0.2 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 | 0.2 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.1 | 1.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 1.8 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 0.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.2 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.1 | 0.4 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.2 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.2 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.1 | 0.1 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.1 | 0.2 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.1 | 0.6 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 | 0.5 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.3 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.5 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.7 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.6 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.3 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.9 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.9 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.3 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.0 | 0.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.9 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.2 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.3 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.0 | GO:0035711 | T-helper 1 cell activation(GO:0035711) |
| 0.0 | 1.2 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.2 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.4 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.2 | GO:2000501 | regulation of natural killer cell chemotaxis(GO:2000501) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 1.0 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.8 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.4 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.7 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.1 | GO:0010266 | response to vitamin B1(GO:0010266) |
| 0.0 | 0.4 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.5 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 1.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.3 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.8 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.0 | 0.4 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.3 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.9 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.2 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.2 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.3 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.3 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.1 | GO:1904747 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.4 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.1 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.1 | GO:0003285 | septum secundum development(GO:0003285) |
| 0.0 | 0.1 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.0 | 0.1 | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.0 | 0.1 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.0 | 0.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.0 | 0.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.7 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.2 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 3.3 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.2 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.3 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.3 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.0 | 0.1 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.2 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.5 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.3 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.2 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.5 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.4 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.3 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.0 | 0.1 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 0.0 | 0.6 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.3 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.3 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.6 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.2 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.0 | GO:0002652 | regulation of tolerance induction dependent upon immune response(GO:0002652) |
| 0.0 | 0.7 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.0 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.2 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.5 | GO:0032527 | protein exit from endoplasmic reticulum(GO:0032527) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.3 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.3 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.5 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.3 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.1 | GO:2001053 | regulation of mesenchymal cell apoptotic process(GO:2001053) negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 0.9 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.4 | GO:0043485 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 | 0.1 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:1904253 | circadian temperature homeostasis(GO:0060086) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.1 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.0 | 0.8 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.0 | 0.2 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 1.6 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.1 | GO:0051532 | regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.1 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.1 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.3 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.1 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.1 | GO:0071503 | response to heparin(GO:0071503) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.8 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 1.3 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.5 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.4 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.4 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.2 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.0 | GO:0015965 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 | 0.1 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) |
| 0.0 | 0.1 | GO:0097354 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.1 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.3 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.0 | 0.0 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 1.3 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.2 | GO:0010561 | negative regulation of glycoprotein biosynthetic process(GO:0010561) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.7 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.1 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.1 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.2 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 3.5 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.0 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.2 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.0 | 0.0 | GO:1900145 | regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.2 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.7 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.4 | GO:1903861 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.0 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.0 | 0.1 | GO:0090070 | regulation of ribosome biogenesis(GO:0090069) positive regulation of ribosome biogenesis(GO:0090070) regulation of rRNA processing(GO:2000232) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0060148 | positive regulation of posttranscriptional gene silencing(GO:0060148) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 0.0 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.3 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.0 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.0 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.4 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.1 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.0 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.1 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.2 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.4 | GO:0055069 | zinc ion homeostasis(GO:0055069) |
| 0.0 | 0.1 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 0.7 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.2 | 0.9 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.2 | 0.6 | GO:0030689 | Noc complex(GO:0030689) |
| 0.2 | 0.6 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.2 | 0.7 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.2 | 0.5 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.4 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 1.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 1.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.8 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 0.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.7 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 0.5 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.3 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 0.3 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.1 | 1.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.5 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.1 | 0.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.4 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.2 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.1 | 0.4 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.5 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 1.7 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 0.5 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.3 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.4 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.1 | 0.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 0.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.6 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.5 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.5 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:1990298 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.0 | 1.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 1.0 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0097134 | Y chromosome(GO:0000806) cyclin A2-CDK2 complex(GO:0097124) cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.3 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.4 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.5 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 3.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 1.2 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.6 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.0 | 0.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.5 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.3 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 1.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.3 | GO:0048500 | signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.0 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 2.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 1.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.2 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.0 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.4 | 1.2 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.3 | 0.8 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.2 | 0.7 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.2 | 0.8 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.2 | 0.8 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 0.9 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.2 | 0.9 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.1 | 0.4 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.4 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.1 | 0.4 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 0.9 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 0.3 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.1 | 0.9 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.6 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.7 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.0 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.3 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.1 | 2.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.3 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.1 | 0.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.4 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.2 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.7 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.4 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.1 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 1.7 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 0.2 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.1 | 0.2 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.1 | 0.2 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.1 | 0.2 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.4 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 0.2 | GO:0038047 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.1 | 0.2 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 0.3 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.4 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 0.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.5 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 1.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 2.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.1 | GO:0098847 | sequence-specific single stranded DNA binding(GO:0098847) |
| 0.1 | 0.6 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 1.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.2 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.4 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.4 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 1.0 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.4 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 2.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 1.7 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 1.0 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.8 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.0 | 0.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.6 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.6 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 2.9 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 1.0 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.2 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.2 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 1.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0052856 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.0 | 0.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.9 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 1.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 0.4 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0001007 | transcription factor activity, RNA polymerase III transcription factor binding(GO:0001007) |
| 0.0 | 0.4 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.2 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.2 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.1 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.8 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 1.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 1.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.7 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.4 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.0 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.0 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 0.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.0 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 1.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.0 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.0 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 0.9 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 1.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 1.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 2.1 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 2.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.7 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.5 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 1.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.5 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.6 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 5.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.2 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 0.8 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 1.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.1 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.0 | 1.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.1 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 3.7 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.5 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.9 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 3.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.9 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 0.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 1.1 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 2.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.4 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.6 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.0 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.8 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.1 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.6 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.4 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |