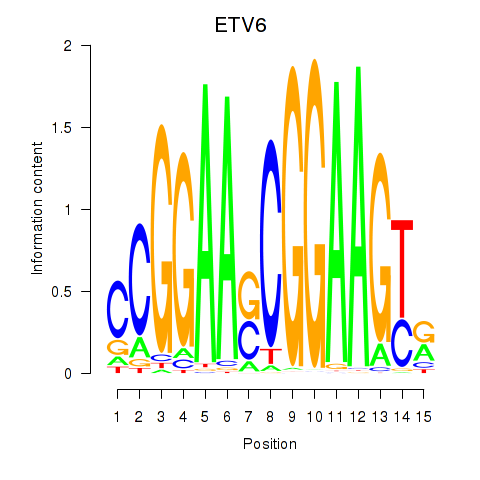
|
chr5_+_137801160
Show fit
|
0.94 |
ENST00000239938.4
|
EGR1
|
early growth response 1
|
|
chr19_+_7069690
Show fit
|
0.94 |
ENST00000439035.2
|
ZNF557
|
zinc finger protein 557
|
|
chr19_+_7069426
Show fit
|
0.85 |
ENST00000252840.6
ENST00000414706.1
|
ZNF557
|
zinc finger protein 557
|
|
chr17_+_77020224
Show fit
|
0.81 |
ENST00000339142.2
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr17_+_77020325
Show fit
|
0.80 |
ENST00000311661.4
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr4_-_47916613
Show fit
|
0.79 |
ENST00000381538.3
ENST00000329043.3
|
NFXL1
|
nuclear transcription factor, X-box binding-like 1
|
|
chr4_-_47916543
Show fit
|
0.76 |
ENST00000507489.1
|
NFXL1
|
nuclear transcription factor, X-box binding-like 1
|
|
chr19_+_39897453
Show fit
|
0.72 |
ENST00000597629.1
ENST00000248673.3
ENST00000594045.1
ENST00000594442.1
|
ZFP36
|
ZFP36 ring finger protein
|
|
chr17_+_77020146
Show fit
|
0.65 |
ENST00000579760.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr19_+_6887571
Show fit
|
0.61 |
ENST00000250572.8
ENST00000381407.5
ENST00000312053.4
ENST00000450315.3
ENST00000381404.4
|
EMR1
|
egf-like module containing, mucin-like, hormone receptor-like 1
|
|
chr12_-_57914275
Show fit
|
0.56 |
ENST00000547303.1
ENST00000552740.1
ENST00000547526.1
ENST00000551116.1
ENST00000346473.3
|
DDIT3
|
DNA-damage-inducible transcript 3
|
|
chr1_+_22351977
Show fit
|
0.56 |
ENST00000420503.1
ENST00000416769.1
ENST00000404210.2
|
LINC00339
|
long intergenic non-protein coding RNA 339
|
|
chr7_+_108210012
Show fit
|
0.51 |
ENST00000249356.3
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9
|
|
chr7_+_155090271
Show fit
|
0.50 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1
|
|
chr7_-_15726296
Show fit
|
0.46 |
ENST00000262041.5
|
MEOX2
|
mesenchyme homeobox 2
|
|
chr5_-_150473127
Show fit
|
0.45 |
ENST00000521001.1
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr14_-_45431091
Show fit
|
0.45 |
ENST00000579157.1
ENST00000396128.4
ENST00000556500.1
|
KLHL28
|
kelch-like family member 28
|
|
chr2_-_27603582
Show fit
|
0.42 |
ENST00000323703.6
ENST00000436006.1
|
ZNF513
|
zinc finger protein 513
|
|
chr9_+_4662282
Show fit
|
0.41 |
ENST00000381883.2
|
PPAPDC2
|
phosphatidic acid phosphatase type 2 domain containing 2
|
|
chr16_+_77224732
Show fit
|
0.41 |
ENST00000569610.1
ENST00000248248.3
ENST00000567291.1
ENST00000320859.6
ENST00000563612.1
ENST00000563279.1
|
MON1B
|
MON1 secretory trafficking family member B
|
|
chr1_-_43855479
Show fit
|
0.38 |
ENST00000290663.6
ENST00000372457.4
|
MED8
|
mediator complex subunit 8
|
|
chr19_-_9929484
Show fit
|
0.37 |
ENST00000586651.1
ENST00000586073.1
|
FBXL12
|
F-box and leucine-rich repeat protein 12
|
|
chr19_-_37663332
Show fit
|
0.37 |
ENST00000392157.2
|
ZNF585A
|
zinc finger protein 585A
|
|
chr6_+_30295036
Show fit
|
0.35 |
ENST00000376659.5
ENST00000428555.1
|
TRIM39
|
tripartite motif containing 39
|
|
chrX_-_125686784
Show fit
|
0.34 |
ENST00000371126.1
|
DCAF12L1
|
DDB1 and CUL4 associated factor 12-like 1
|
|
chr19_-_37663572
Show fit
|
0.34 |
ENST00000588354.1
ENST00000292841.5
ENST00000355533.2
ENST00000356958.4
|
ZNF585A
|
zinc finger protein 585A
|
|
chr19_-_9929708
Show fit
|
0.34 |
ENST00000247977.4
ENST00000590277.1
ENST00000588922.1
ENST00000589626.1
ENST00000592067.1
ENST00000586469.1
|
FBXL12
|
F-box and leucine-rich repeat protein 12
|
|
chr9_-_100684845
Show fit
|
0.33 |
ENST00000375119.3
|
C9orf156
|
chromosome 9 open reading frame 156
|
|
chr12_-_48551366
Show fit
|
0.32 |
ENST00000535988.1
ENST00000536953.1
ENST00000535055.1
ENST00000317697.3
ENST00000536549.1
|
ASB8
|
ankyrin repeat and SOCS box containing 8
|
|
chr17_-_33905521
Show fit
|
0.32 |
ENST00000225873.4
|
PEX12
|
peroxisomal biogenesis factor 12
|
|
chr14_+_24616588
Show fit
|
0.32 |
ENST00000324103.6
ENST00000559260.1
|
RNF31
|
ring finger protein 31
|
|
chr6_+_30525051
Show fit
|
0.31 |
ENST00000376557.3
|
PRR3
|
proline rich 3
|
|
chr10_-_79789291
Show fit
|
0.31 |
ENST00000372371.3
|
POLR3A
|
polymerase (RNA) III (DNA directed) polypeptide A, 155kDa
|
|
chr6_+_30524663
Show fit
|
0.31 |
ENST00000376560.3
|
PRR3
|
proline rich 3
|
|
chr16_-_23652570
Show fit
|
0.30 |
ENST00000261584.4
|
PALB2
|
partner and localizer of BRCA2
|
|
chr19_-_1479532
Show fit
|
0.30 |
ENST00000436106.2
|
C19orf25
|
chromosome 19 open reading frame 25
|
|
chr6_-_28367510
Show fit
|
0.30 |
ENST00000361028.1
|
ZSCAN12
|
zinc finger and SCAN domain containing 12
|
|
chr19_-_1479117
Show fit
|
0.29 |
ENST00000586564.1
ENST00000589529.1
ENST00000585675.1
ENST00000592872.1
ENST00000588871.1
ENST00000588427.1
ENST00000427685.2
|
C19orf25
|
chromosome 19 open reading frame 25
|
|
chr9_-_95640218
Show fit
|
0.29 |
ENST00000395506.3
ENST00000375495.3
ENST00000332591.6
|
ZNF484
|
zinc finger protein 484
|
|
chr12_+_120105558
Show fit
|
0.29 |
ENST00000229328.5
ENST00000541640.1
|
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit
|
|
chr17_+_79650962
Show fit
|
0.29 |
ENST00000329138.4
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr16_+_3355472
Show fit
|
0.29 |
ENST00000574298.1
|
ZNF75A
|
zinc finger protein 75a
|
|
chr6_+_30294612
Show fit
|
0.29 |
ENST00000440271.1
ENST00000396551.3
ENST00000376656.4
ENST00000540416.1
ENST00000428728.1
ENST00000396548.1
ENST00000428404.1
|
TRIM39
|
tripartite motif containing 39
|
|
chr16_+_3493611
Show fit
|
0.29 |
ENST00000407558.4
ENST00000572169.1
ENST00000572757.1
ENST00000573593.1
ENST00000570372.1
ENST00000424546.2
ENST00000575733.1
ENST00000573201.1
ENST00000574950.1
ENST00000573580.1
ENST00000608722.1
|
NAA60
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit
N-alpha-acetyltransferase 60
|
|
chr2_-_219537134
Show fit
|
0.28 |
ENST00000295704.2
|
RNF25
|
ring finger protein 25
|
|
chr7_+_148823485
Show fit
|
0.28 |
ENST00000426851.2
|
ZNF398
|
zinc finger protein 398
|
|
chr1_-_55266865
Show fit
|
0.28 |
ENST00000371274.4
|
TTC22
|
tetratricopeptide repeat domain 22
|
|
chrX_+_47092314
Show fit
|
0.28 |
ENST00000218348.3
|
USP11
|
ubiquitin specific peptidase 11
|
|
chr2_+_220462560
Show fit
|
0.28 |
ENST00000456909.1
ENST00000295641.10
|
STK11IP
|
serine/threonine kinase 11 interacting protein
|
|
chr14_-_100842588
Show fit
|
0.28 |
ENST00000556645.1
ENST00000556209.1
ENST00000556504.1
ENST00000556435.1
ENST00000554772.1
ENST00000553581.1
ENST00000553769.2
ENST00000554605.1
ENST00000557722.1
ENST00000553413.1
ENST00000553524.1
ENST00000358655.4
|
WARS
|
tryptophanyl-tRNA synthetase
|
|
chr11_+_63993738
Show fit
|
0.27 |
ENST00000441250.2
ENST00000279206.3
|
NUDT22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22
|
|
chr19_-_37701386
Show fit
|
0.27 |
ENST00000527838.1
ENST00000591492.1
ENST00000532828.2
|
ZNF585B
|
zinc finger protein 585B
|
|
chr14_-_24616426
Show fit
|
0.27 |
ENST00000216802.5
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta)
|
|
chr19_+_45681997
Show fit
|
0.27 |
ENST00000433642.2
|
BLOC1S3
|
biogenesis of lysosomal organelles complex-1, subunit 3
|
|
chr1_+_24069642
Show fit
|
0.27 |
ENST00000418390.2
|
TCEB3
|
transcription elongation factor B (SIII), polypeptide 3 (110kDa, elongin A)
|
|
chr12_-_6798523
Show fit
|
0.26 |
ENST00000319770.3
|
ZNF384
|
zinc finger protein 384
|
|
chr16_+_1662326
Show fit
|
0.26 |
ENST00000397412.3
|
CRAMP1L
|
Crm, cramped-like (Drosophila)
|
|
chr12_-_6798410
Show fit
|
0.26 |
ENST00000361959.3
ENST00000436774.2
ENST00000544482.1
|
ZNF384
|
zinc finger protein 384
|
|
chr10_+_99079008
Show fit
|
0.26 |
ENST00000371021.3
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr20_+_18269121
Show fit
|
0.26 |
ENST00000377671.3
ENST00000360010.5
ENST00000396026.3
ENST00000402618.2
ENST00000401790.1
ENST00000434018.1
ENST00000538547.1
ENST00000535822.1
|
ZNF133
|
zinc finger protein 133
|
|
chr9_-_140095186
Show fit
|
0.26 |
ENST00000409012.4
|
TPRN
|
taperin
|
|
chr17_-_79876010
Show fit
|
0.26 |
ENST00000328666.6
|
SIRT7
|
sirtuin 7
|
|
chr16_+_3507985
Show fit
|
0.25 |
ENST00000421765.3
ENST00000360862.5
ENST00000414063.2
ENST00000610180.1
ENST00000608993.1
|
NAA60
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit
N-alpha-acetyltransferase 60
|
|
chr8_-_145980968
Show fit
|
0.25 |
ENST00000292562.7
|
ZNF251
|
zinc finger protein 251
|
|
chrX_+_122993657
Show fit
|
0.25 |
ENST00000434753.3
ENST00000430625.1
|
XIAP
|
X-linked inhibitor of apoptosis
|
|
chr11_-_118122996
Show fit
|
0.25 |
ENST00000525386.1
ENST00000527472.1
ENST00000278949.4
|
MPZL3
|
myelin protein zero-like 3
|
|
chr7_-_99756293
Show fit
|
0.25 |
ENST00000316937.3
ENST00000456769.1
|
C7orf43
|
chromosome 7 open reading frame 43
|
|
chr14_+_45431379
Show fit
|
0.25 |
ENST00000361577.3
ENST00000361462.2
ENST00000382233.2
|
FAM179B
|
family with sequence similarity 179, member B
|
|
chr12_-_6798616
Show fit
|
0.25 |
ENST00000355772.4
ENST00000417772.3
ENST00000396801.3
ENST00000396799.2
|
ZNF384
|
zinc finger protein 384
|
|
chr17_-_76836729
Show fit
|
0.25 |
ENST00000587783.1
ENST00000542802.3
ENST00000586531.1
ENST00000589424.1
ENST00000590546.2
|
USP36
|
ubiquitin specific peptidase 36
|
|
chr3_+_38080691
Show fit
|
0.25 |
ENST00000308059.6
ENST00000346219.3
ENST00000452631.2
|
DLEC1
|
deleted in lung and esophageal cancer 1
|
|
chr1_-_38019878
Show fit
|
0.25 |
ENST00000296215.6
|
SNIP1
|
Smad nuclear interacting protein 1
|
|
chr3_-_52740012
Show fit
|
0.24 |
ENST00000407584.3
ENST00000266014.5
|
GLT8D1
|
glycosyltransferase 8 domain containing 1
|
|
chr15_+_75074385
Show fit
|
0.24 |
ENST00000220003.9
|
CSK
|
c-src tyrosine kinase
|
|
chr22_+_37309662
Show fit
|
0.24 |
ENST00000403662.3
ENST00000262825.5
|
CSF2RB
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage)
|
|
chr3_-_52739762
Show fit
|
0.24 |
ENST00000487642.1
ENST00000464705.1
ENST00000491606.1
ENST00000489119.1
ENST00000478968.2
|
GLT8D1
|
glycosyltransferase 8 domain containing 1
|
|
chr3_-_16555150
Show fit
|
0.24 |
ENST00000334133.4
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr19_+_44576296
Show fit
|
0.24 |
ENST00000421176.3
|
ZNF284
|
zinc finger protein 284
|
|
chrX_+_48398053
Show fit
|
0.23 |
ENST00000537536.1
ENST00000418627.1
|
TBC1D25
|
TBC1 domain family, member 25
|
|
chr22_-_19166343
Show fit
|
0.23 |
ENST00000215882.5
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr16_-_3493528
Show fit
|
0.23 |
ENST00000301744.4
|
ZNF597
|
zinc finger protein 597
|
|
chr19_+_11670245
Show fit
|
0.23 |
ENST00000585493.1
|
ZNF627
|
zinc finger protein 627
|
|
chr17_-_76836963
Show fit
|
0.23 |
ENST00000312010.6
|
USP36
|
ubiquitin specific peptidase 36
|
|
chr1_+_24069952
Show fit
|
0.23 |
ENST00000609199.1
|
TCEB3
|
transcription elongation factor B (SIII), polypeptide 3 (110kDa, elongin A)
|
|
chr12_+_124196865
Show fit
|
0.23 |
ENST00000330342.3
|
ATP6V0A2
|
ATPase, H+ transporting, lysosomal V0 subunit a2
|
|
chr11_-_207221
Show fit
|
0.22 |
ENST00000486280.1
ENST00000332865.6
ENST00000529614.2
ENST00000325147.9
ENST00000410108.1
ENST00000382762.3
|
BET1L
|
Bet1 golgi vesicular membrane trafficking protein-like
|
|
chr17_+_75137460
Show fit
|
0.22 |
ENST00000587820.1
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae)
|
|
chr21_-_30365136
Show fit
|
0.22 |
ENST00000361371.5
ENST00000389194.2
ENST00000389195.2
|
LTN1
|
listerin E3 ubiquitin protein ligase 1
|
|
chr6_-_28367481
Show fit
|
0.22 |
ENST00000396827.3
|
ZSCAN12
|
zinc finger and SCAN domain containing 12
|
|
chr1_-_246729544
Show fit
|
0.22 |
ENST00000544618.1
ENST00000366514.4
|
TFB2M
|
transcription factor B2, mitochondrial
|
|
chr11_-_71814422
Show fit
|
0.22 |
ENST00000278671.5
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr9_-_131709858
Show fit
|
0.22 |
ENST00000372586.3
|
DOLK
|
dolichol kinase
|
|
chr5_+_177557997
Show fit
|
0.22 |
ENST00000313386.4
ENST00000515098.1
ENST00000542098.1
ENST00000502814.1
ENST00000507457.1
ENST00000508647.1
|
RMND5B
|
required for meiotic nuclear division 5 homolog B (S. cerevisiae)
|
|
chr19_-_16682987
Show fit
|
0.22 |
ENST00000431408.1
ENST00000436553.2
ENST00000595753.1
|
SLC35E1
|
solute carrier family 35, member E1
|
|
chr2_+_3383439
Show fit
|
0.21 |
ENST00000382110.2
ENST00000324266.5
|
TRAPPC12
|
trafficking protein particle complex 12
|
|
chr17_+_5389605
Show fit
|
0.21 |
ENST00000576988.1
ENST00000576570.1
ENST00000573759.1
|
MIS12
|
MIS12 kinetochore complex component
|
|
chr6_+_170615819
Show fit
|
0.21 |
ENST00000476287.1
ENST00000252510.9
|
FAM120B
|
family with sequence similarity 120B
|
|
chr1_-_55266926
Show fit
|
0.21 |
ENST00000371276.4
|
TTC22
|
tetratricopeptide repeat domain 22
|
|
chr20_+_33464407
Show fit
|
0.21 |
ENST00000253382.5
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2
|
|
chr11_-_47600549
Show fit
|
0.21 |
ENST00000430070.2
|
KBTBD4
|
kelch repeat and BTB (POZ) domain containing 4
|
|
chr17_-_61523535
Show fit
|
0.21 |
ENST00000584031.1
ENST00000392976.1
|
CYB561
|
cytochrome b561
|
|
chr15_-_65809581
Show fit
|
0.21 |
ENST00000341861.5
|
DPP8
|
dipeptidyl-peptidase 8
|
|
chr18_-_47813940
Show fit
|
0.20 |
ENST00000586837.1
ENST00000412036.2
ENST00000589940.1
|
CXXC1
|
CXXC finger protein 1
|
|
chr20_+_33464238
Show fit
|
0.20 |
ENST00000360596.2
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2
|
|
chr16_+_67226019
Show fit
|
0.20 |
ENST00000379378.3
|
E2F4
|
E2F transcription factor 4, p107/p130-binding
|
|
chr6_+_43603552
Show fit
|
0.20 |
ENST00000372171.4
|
MAD2L1BP
|
MAD2L1 binding protein
|
|
chrX_+_27826107
Show fit
|
0.20 |
ENST00000356790.2
|
MAGEB10
|
melanoma antigen family B, 10
|
|
chr19_+_17186577
Show fit
|
0.20 |
ENST00000595618.1
ENST00000594824.1
|
MYO9B
|
myosin IXB
|
|
chr10_-_99094458
Show fit
|
0.20 |
ENST00000371019.2
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2
|
|
chr11_-_47600320
Show fit
|
0.20 |
ENST00000525720.1
ENST00000531067.1
ENST00000533290.1
ENST00000529499.1
ENST00000529946.1
ENST00000526005.1
ENST00000395288.2
ENST00000534239.1
|
KBTBD4
|
kelch repeat and BTB (POZ) domain containing 4
|
|
chr11_+_68671310
Show fit
|
0.20 |
ENST00000255078.3
ENST00000539224.1
|
IGHMBP2
|
immunoglobulin mu binding protein 2
|
|
chr17_+_18601299
Show fit
|
0.20 |
ENST00000572555.1
ENST00000395902.3
ENST00000449552.2
|
TRIM16L
|
tripartite motif containing 16-like
|
|
chr17_-_5389477
Show fit
|
0.20 |
ENST00000572834.1
ENST00000570848.1
ENST00000571971.1
ENST00000158771.4
|
DERL2
|
derlin 2
|
|
chr1_-_20834586
Show fit
|
0.20 |
ENST00000264198.3
|
MUL1
|
mitochondrial E3 ubiquitin protein ligase 1
|
|
chr1_+_38273419
Show fit
|
0.19 |
ENST00000468084.1
|
C1orf122
|
chromosome 1 open reading frame 122
|
|
chr12_+_54402790
Show fit
|
0.19 |
ENST00000040584.4
|
HOXC8
|
homeobox C8
|
|
chr12_-_125348329
Show fit
|
0.19 |
ENST00000546215.1
ENST00000415380.2
ENST00000261693.6
ENST00000376788.1
ENST00000545493.1
|
SCARB1
|
scavenger receptor class B, member 1
|
|
chr16_+_31085714
Show fit
|
0.19 |
ENST00000300850.5
ENST00000564189.1
ENST00000428260.1
|
ZNF646
|
zinc finger protein 646
|
|
chr19_+_16308711
Show fit
|
0.19 |
ENST00000429941.2
ENST00000444449.2
ENST00000589822.1
|
AP1M1
|
adaptor-related protein complex 1, mu 1 subunit
|
|
chr19_+_16308659
Show fit
|
0.19 |
ENST00000590263.1
ENST00000590756.1
ENST00000541844.1
|
AP1M1
|
adaptor-related protein complex 1, mu 1 subunit
|
|
chr7_+_150929550
Show fit
|
0.19 |
ENST00000482173.1
ENST00000495645.1
ENST00000035307.2
|
CHPF2
|
chondroitin polymerizing factor 2
|
|
chr4_-_5710257
Show fit
|
0.18 |
ENST00000344938.1
ENST00000344408.5
|
EVC2
|
Ellis van Creveld syndrome 2
|
|
chr16_+_15737124
Show fit
|
0.18 |
ENST00000396355.1
ENST00000396353.2
|
NDE1
|
nudE neurodevelopment protein 1
|
|
chr17_-_61523622
Show fit
|
0.18 |
ENST00000448884.2
ENST00000582297.1
ENST00000582034.1
ENST00000578072.1
ENST00000360793.3
|
CYB561
|
cytochrome b561
|
|
chr1_+_230883179
Show fit
|
0.18 |
ENST00000366666.2
|
CAPN9
|
calpain 9
|
|
chr1_+_156163880
Show fit
|
0.18 |
ENST00000359511.4
ENST00000423538.2
|
SLC25A44
|
solute carrier family 25, member 44
|
|
chr16_+_31470143
Show fit
|
0.18 |
ENST00000457010.2
ENST00000563544.1
|
ARMC5
|
armadillo repeat containing 5
|
|
chr17_-_79650689
Show fit
|
0.18 |
ENST00000574938.1
ENST00000570561.1
ENST00000573392.1
ENST00000576135.1
|
ARL16
|
ADP-ribosylation factor-like 16
|
|
chr1_-_38325256
Show fit
|
0.18 |
ENST00000373036.4
|
MTF1
|
metal-regulatory transcription factor 1
|
|
chr7_+_44240520
Show fit
|
0.18 |
ENST00000496112.1
ENST00000223369.2
|
YKT6
|
YKT6 v-SNARE homolog (S. cerevisiae)
|
|
chr9_+_100818976
Show fit
|
0.17 |
ENST00000210444.5
|
NANS
|
N-acetylneuraminic acid synthase
|
|
chr16_-_28074822
Show fit
|
0.17 |
ENST00000395724.3
ENST00000380898.2
ENST00000447459.2
|
GSG1L
|
GSG1-like
|
|
chr17_-_79650818
Show fit
|
0.17 |
ENST00000397498.4
|
ARL16
|
ADP-ribosylation factor-like 16
|
|
chr16_+_3508063
Show fit
|
0.17 |
ENST00000576787.1
ENST00000572942.1
ENST00000576916.1
ENST00000575076.1
ENST00000572131.1
|
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit
|
|
chr11_+_207477
Show fit
|
0.17 |
ENST00000526104.1
|
RIC8A
|
RIC8 guanine nucleotide exchange factor A
|
|
chr9_+_21967137
Show fit
|
0.17 |
ENST00000441769.2
|
C9orf53
|
chromosome 9 open reading frame 53
|
|
chr4_+_492985
Show fit
|
0.17 |
ENST00000296306.7
ENST00000536264.1
ENST00000310340.5
ENST00000453061.2
ENST00000504346.1
ENST00000503111.1
ENST00000383028.4
ENST00000509768.1
|
PIGG
|
phosphatidylinositol glycan anchor biosynthesis, class G
|
|
chr11_+_695787
Show fit
|
0.17 |
ENST00000526170.1
ENST00000488769.1
|
TMEM80
|
transmembrane protein 80
|
|
chr19_-_44952635
Show fit
|
0.17 |
ENST00000592308.1
ENST00000588931.1
ENST00000291187.4
|
ZNF229
|
zinc finger protein 229
|
|
chr16_+_31470179
Show fit
|
0.17 |
ENST00000538189.1
ENST00000268314.4
|
ARMC5
|
armadillo repeat containing 5
|
|
chr19_-_9731872
Show fit
|
0.17 |
ENST00000424629.1
ENST00000326044.5
ENST00000354661.4
ENST00000435550.1
ENST00000444611.1
ENST00000421525.1
|
ZNF561
|
zinc finger protein 561
|
|
chr6_-_31633402
Show fit
|
0.17 |
ENST00000375893.2
|
GPANK1
|
G patch domain and ankyrin repeats 1
|
|
chr10_-_22292613
Show fit
|
0.17 |
ENST00000376980.3
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1
|
|
chr16_+_2205755
Show fit
|
0.17 |
ENST00000326181.6
|
TRAF7
|
TNF receptor-associated factor 7, E3 ubiquitin protein ligase
|
|
chr8_+_125985531
Show fit
|
0.17 |
ENST00000319286.5
|
ZNF572
|
zinc finger protein 572
|
|
chr4_+_980785
Show fit
|
0.17 |
ENST00000247933.4
ENST00000453894.1
|
IDUA
|
iduronidase, alpha-L-
|
|
chr19_-_9546177
Show fit
|
0.16 |
ENST00000592292.1
ENST00000588221.1
|
ZNF266
|
zinc finger protein 266
|
|
chr9_+_127631399
Show fit
|
0.16 |
ENST00000259477.6
|
ARPC5L
|
actin related protein 2/3 complex, subunit 5-like
|
|
chrX_-_48768913
Show fit
|
0.16 |
ENST00000376529.3
ENST00000247138.5
ENST00000413561.2
ENST00000452555.2
ENST00000445167.2
ENST00000376512.1
|
SLC35A2
|
solute carrier family 35 (UDP-galactose transporter), member A2
|
|
chr12_-_76742183
Show fit
|
0.16 |
ENST00000393262.3
|
BBS10
|
Bardet-Biedl syndrome 10
|
|
chr12_+_54447637
Show fit
|
0.16 |
ENST00000609810.1
ENST00000430889.2
|
HOXC4
HOXC4
|
homeobox C4
Homeobox protein Hox-C4
|
|
chr9_-_34126730
Show fit
|
0.16 |
ENST00000361264.4
|
DCAF12
|
DDB1 and CUL4 associated factor 12
|
|
chr1_-_43855444
Show fit
|
0.16 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8
|
|
chr11_-_61129723
Show fit
|
0.16 |
ENST00000537680.1
ENST00000426130.2
ENST00000294072.4
|
CYB561A3
|
cytochrome b561 family, member A3
|
|
chr22_+_38004832
Show fit
|
0.16 |
ENST00000405147.3
ENST00000429218.1
ENST00000325180.8
ENST00000337437.4
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1
|
|
chr11_-_123612319
Show fit
|
0.16 |
ENST00000526252.1
ENST00000530393.1
ENST00000533463.1
ENST00000336139.4
ENST00000529691.1
ENST00000528306.1
|
ZNF202
|
zinc finger protein 202
|
|
chr1_+_22979474
Show fit
|
0.16 |
ENST00000509305.1
|
C1QB
|
complement component 1, q subcomponent, B chain
|
|
chr3_+_127771212
Show fit
|
0.16 |
ENST00000243253.3
ENST00000481210.1
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|
|
chr17_+_19030782
Show fit
|
0.16 |
ENST00000344415.4
ENST00000577213.1
|
GRAPL
|
GRB2-related adaptor protein-like
|
|
chr16_-_69373396
Show fit
|
0.15 |
ENST00000562595.1
ENST00000562081.1
ENST00000306875.4
|
COG8
|
component of oligomeric golgi complex 8
|
|
chr14_+_96000930
Show fit
|
0.15 |
ENST00000331334.4
|
GLRX5
|
glutaredoxin 5
|
|
chrX_+_51546103
Show fit
|
0.15 |
ENST00000375772.3
|
MAGED1
|
melanoma antigen family D, 1
|
|
chr15_-_80263506
Show fit
|
0.15 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1
|
|
chr10_+_75504105
Show fit
|
0.15 |
ENST00000535742.1
ENST00000546025.1
ENST00000345254.4
ENST00000540668.1
ENST00000339365.2
ENST00000411652.2
|
SEC24C
|
SEC24 family member C
|
|
chr20_-_3140490
Show fit
|
0.15 |
ENST00000449731.1
ENST00000380266.3
|
UBOX5
FASTKD5
|
U-box domain containing 5
FAST kinase domains 5
|
|
chr1_+_27648709
Show fit
|
0.15 |
ENST00000608611.1
ENST00000466759.1
ENST00000464813.1
ENST00000498220.1
|
TMEM222
|
transmembrane protein 222
|
|
chr11_-_65769594
Show fit
|
0.15 |
ENST00000532707.1
ENST00000533544.1
ENST00000526451.1
ENST00000312234.2
ENST00000530462.1
ENST00000525767.1
ENST00000529964.1
ENST00000527249.1
|
EIF1AD
|
eukaryotic translation initiation factor 1A domain containing
|
|
chr19_-_37263693
Show fit
|
0.15 |
ENST00000591344.1
|
ZNF850
|
zinc finger protein 850
|
|
chr1_+_27648648
Show fit
|
0.15 |
ENST00000374076.4
|
TMEM222
|
transmembrane protein 222
|
|
chr15_-_41836441
Show fit
|
0.15 |
ENST00000567866.1
ENST00000561603.1
ENST00000304330.4
ENST00000566863.1
|
RPAP1
|
RNA polymerase II associated protein 1
|
|
chr6_-_108395907
Show fit
|
0.15 |
ENST00000193322.3
|
OSTM1
|
osteopetrosis associated transmembrane protein 1
|
|
chr12_+_50478977
Show fit
|
0.15 |
ENST00000381513.4
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1
|
|
chr2_+_96931834
Show fit
|
0.15 |
ENST00000488633.1
|
CIAO1
|
cytosolic iron-sulfur protein assembly 1
|
|
chr1_+_38273818
Show fit
|
0.15 |
ENST00000373042.4
|
C1orf122
|
chromosome 1 open reading frame 122
|
|
chr19_+_56186557
Show fit
|
0.15 |
ENST00000270460.6
|
EPN1
|
epsin 1
|
|
chr3_+_40518599
Show fit
|
0.15 |
ENST00000314686.5
ENST00000447116.2
ENST00000429348.2
ENST00000456778.1
|
ZNF619
|
zinc finger protein 619
|
|
chr19_-_45737469
Show fit
|
0.15 |
ENST00000413988.1
|
EXOC3L2
|
exocyst complex component 3-like 2
|
|
chr17_-_18950310
Show fit
|
0.15 |
ENST00000573099.1
|
GRAP
|
GRB2-related adaptor protein
|
|
chr11_+_65479462
Show fit
|
0.15 |
ENST00000377046.3
ENST00000352980.4
ENST00000341318.4
|
KAT5
|
K(lysine) acetyltransferase 5
|
|
chr16_+_27561449
Show fit
|
0.15 |
ENST00000261588.4
|
KIAA0556
|
KIAA0556
|
|
chr11_+_65479702
Show fit
|
0.15 |
ENST00000530446.1
ENST00000534104.1
ENST00000530605.1
ENST00000528198.1
ENST00000531880.1
ENST00000534650.1
|
KAT5
|
K(lysine) acetyltransferase 5
|
|
chr1_+_43637996
Show fit
|
0.15 |
ENST00000528956.1
ENST00000529956.1
|
WDR65
|
WD repeat domain 65
|
|
chr11_+_57435219
Show fit
|
0.15 |
ENST00000527985.1
ENST00000287169.3
|
ZDHHC5
|
zinc finger, DHHC-type containing 5
|
|
chr17_-_56595196
Show fit
|
0.14 |
ENST00000579921.1
ENST00000579925.1
ENST00000323456.5
|
MTMR4
|
myotubularin related protein 4
|
|
chr11_-_71814276
Show fit
|
0.14 |
ENST00000538404.1
ENST00000535107.1
ENST00000545249.1
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr15_-_51058005
Show fit
|
0.14 |
ENST00000261854.5
|
SPPL2A
|
signal peptide peptidase like 2A
|
|
chr1_-_1209106
Show fit
|
0.14 |
ENST00000360466.2
ENST00000509720.1
ENST00000400930.4
ENST00000422076.1
ENST00000435198.1
ENST00000347370.2
ENST00000502382.1
ENST00000400929.2
ENST00000348298.7
ENST00000349431.6
|
UBE2J2
|
ubiquitin-conjugating enzyme E2, J2
|
|
chr13_+_29233218
Show fit
|
0.14 |
ENST00000380842.4
|
POMP
|
proteasome maturation protein
|
|
chr12_+_64845660
Show fit
|
0.14 |
ENST00000331710.5
|
TBK1
|
TANK-binding kinase 1
|
|
chr5_-_6378700
Show fit
|
0.14 |
ENST00000255764.3
|
MED10
|
mediator complex subunit 10
|
|
chr18_-_45457192
Show fit
|
0.14 |
ENST00000586514.1
ENST00000591214.1
ENST00000589877.1
|
SMAD2
|
SMAD family member 2
|
|
chr19_-_44860820
Show fit
|
0.14 |
ENST00000354340.4
ENST00000337401.4
ENST00000587909.1
|
ZNF112
|
zinc finger protein 112
|
|
chrX_-_107018969
Show fit
|
0.14 |
ENST00000372383.4
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr5_-_102455801
Show fit
|
0.14 |
ENST00000508629.1
ENST00000399004.2
|
GIN1
|
gypsy retrotransposon integrase 1
|
|
chr22_+_38005033
Show fit
|
0.14 |
ENST00000447515.1
ENST00000406772.1
ENST00000431745.1
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1
|
|
chr1_-_114301755
Show fit
|
0.14 |
ENST00000393357.2
ENST00000369596.2
ENST00000446739.1
|
PHTF1
|
putative homeodomain transcription factor 1
|
|
chrX_+_46306624
Show fit
|
0.14 |
ENST00000360017.5
|
KRBOX4
|
KRAB box domain containing 4
|
|
chrX_+_1710484
Show fit
|
0.14 |
ENST00000313871.3
ENST00000381261.3
|
AKAP17A
|
A kinase (PRKA) anchor protein 17A
|
|
chr19_+_45542773
Show fit
|
0.14 |
ENST00000544944.2
|
CLASRP
|
CLK4-associating serine/arginine rich protein
|
|
chr19_+_1249869
Show fit
|
0.14 |
ENST00000591446.2
|
MIDN
|
midnolin
|