Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
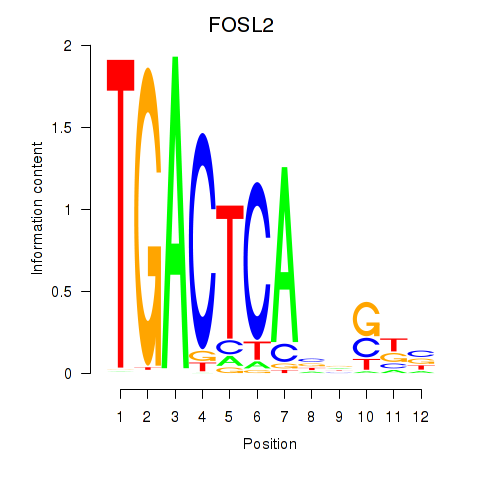
Results for FOSL2_SMARCC1
Z-value: 0.79
Transcription factors associated with FOSL2_SMARCC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOSL2
|
ENSG00000075426.7 | FOS like 2, AP-1 transcription factor subunit |
|
SMARCC1
|
ENSG00000173473.6 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOSL2 | hg19_v2_chr2_+_28615669_28615733 | 0.34 | 9.3e-02 | Click! |
| SMARCC1 | hg19_v2_chr3_-_47823298_47823423 | 0.27 | 1.9e-01 | Click! |
Activity profile of FOSL2_SMARCC1 motif
Sorted Z-values of FOSL2_SMARCC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_74606223 | 3.58 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr7_+_30960915 | 2.87 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr12_-_49259643 | 2.37 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr13_+_32838801 | 2.13 |
ENST00000542859.1
|
FRY
|
furry homolog (Drosophila) |
| chr15_+_67420441 | 2.05 |
ENST00000558894.1
|
SMAD3
|
SMAD family member 3 |
| chr15_+_67458357 | 1.83 |
ENST00000537194.2
|
SMAD3
|
SMAD family member 3 |
| chr10_+_104155450 | 1.81 |
ENST00000471698.1
ENST00000189444.6 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr21_+_26934165 | 1.73 |
ENST00000456917.1
|
MIR155HG
|
MIR155 host gene (non-protein coding) |
| chr3_+_11178779 | 1.71 |
ENST00000438284.2
|
HRH1
|
histamine receptor H1 |
| chr15_+_67418047 | 1.43 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr7_+_129932974 | 1.43 |
ENST00000445470.2
ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chr2_+_169926047 | 1.32 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr5_-_176923846 | 1.24 |
ENST00000506537.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr18_+_21452804 | 1.16 |
ENST00000269217.6
|
LAMA3
|
laminin, alpha 3 |
| chr5_-_176923803 | 1.13 |
ENST00000506161.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr7_+_150065278 | 1.10 |
ENST00000519397.1
ENST00000479668.1 ENST00000540729.1 |
REPIN1
|
replication initiator 1 |
| chr4_+_156587979 | 1.09 |
ENST00000511507.1
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr22_+_23046750 | 1.05 |
ENST00000390307.2
|
IGLV3-22
|
immunoglobulin lambda variable 3-22 (gene/pseudogene) |
| chr4_+_108745711 | 1.04 |
ENST00000394684.4
|
SGMS2
|
sphingomyelin synthase 2 |
| chr16_-_4665023 | 0.96 |
ENST00000591897.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr2_+_201980827 | 0.94 |
ENST00000309955.3
ENST00000443227.1 ENST00000341222.6 ENST00000355558.4 ENST00000340870.5 ENST00000341582.6 |
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr2_+_102721023 | 0.88 |
ENST00000409589.1
ENST00000409329.1 |
IL1R1
|
interleukin 1 receptor, type I |
| chr16_-_4664860 | 0.88 |
ENST00000587615.1
ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1
|
UBA-like domain containing 1 |
| chr7_+_73245193 | 0.85 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr2_+_152214098 | 0.84 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr17_+_21191341 | 0.83 |
ENST00000526076.2
ENST00000361818.5 ENST00000316920.6 |
MAP2K3
|
mitogen-activated protein kinase kinase 3 |
| chr11_+_35639735 | 0.82 |
ENST00000317811.4
|
FJX1
|
four jointed box 1 (Drosophila) |
| chr3_+_184037466 | 0.82 |
ENST00000441154.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr2_+_201981527 | 0.81 |
ENST00000441224.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr2_+_169923577 | 0.77 |
ENST00000432060.2
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr7_+_48128194 | 0.76 |
ENST00000416681.1
ENST00000331803.4 ENST00000432131.1 |
UPP1
|
uridine phosphorylase 1 |
| chr5_+_150020214 | 0.76 |
ENST00000307662.4
|
SYNPO
|
synaptopodin |
| chr7_+_48128316 | 0.76 |
ENST00000341253.4
|
UPP1
|
uridine phosphorylase 1 |
| chr1_-_6662919 | 0.75 |
ENST00000377658.4
ENST00000377663.3 |
KLHL21
|
kelch-like family member 21 |
| chr17_+_7255208 | 0.73 |
ENST00000333751.3
|
KCTD11
|
potassium channel tetramerization domain containing 11 |
| chr21_-_35899113 | 0.73 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr6_-_44233361 | 0.69 |
ENST00000275015.5
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr3_+_124223586 | 0.69 |
ENST00000393496.1
|
KALRN
|
kalirin, RhoGEF kinase |
| chr3_-_149293990 | 0.69 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr7_+_94023873 | 0.69 |
ENST00000297268.6
|
COL1A2
|
collagen, type I, alpha 2 |
| chr6_-_159239257 | 0.67 |
ENST00000337147.7
ENST00000392177.4 |
EZR
|
ezrin |
| chr2_+_36923830 | 0.65 |
ENST00000379242.3
ENST00000389975.3 |
VIT
|
vitrin |
| chr15_+_89182178 | 0.64 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr1_-_89488510 | 0.64 |
ENST00000564665.1
ENST00000370481.4 |
GBP3
|
guanylate binding protein 3 |
| chr6_+_138188551 | 0.63 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr21_+_17961006 | 0.62 |
ENST00000602323.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr16_+_58533951 | 0.62 |
ENST00000566192.1
ENST00000565088.1 ENST00000568640.1 ENST00000563978.1 ENST00000569923.1 ENST00000356752.4 ENST00000563799.1 ENST00000562999.1 ENST00000570248.1 ENST00000562731.1 ENST00000568424.1 |
NDRG4
|
NDRG family member 4 |
| chr2_+_169923504 | 0.61 |
ENST00000357546.2
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr11_+_101983176 | 0.61 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr6_-_24877490 | 0.60 |
ENST00000540914.1
ENST00000378023.4 |
FAM65B
|
family with sequence similarity 65, member B |
| chr10_+_75670862 | 0.60 |
ENST00000446342.1
ENST00000372764.3 ENST00000372762.4 |
PLAU
|
plasminogen activator, urokinase |
| chr12_-_56236734 | 0.59 |
ENST00000548629.1
|
MMP19
|
matrix metallopeptidase 19 |
| chr15_+_59903975 | 0.59 |
ENST00000560585.1
ENST00000396065.1 |
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr15_+_89181974 | 0.58 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr9_-_74979420 | 0.58 |
ENST00000343431.2
ENST00000376956.3 |
ZFAND5
|
zinc finger, AN1-type domain 5 |
| chr8_-_141774467 | 0.58 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr1_-_160231451 | 0.55 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr6_+_106546808 | 0.55 |
ENST00000369089.3
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr2_-_145275228 | 0.55 |
ENST00000427902.1
ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr12_+_100660909 | 0.55 |
ENST00000549687.1
|
SCYL2
|
SCY1-like 2 (S. cerevisiae) |
| chr15_+_89182156 | 0.54 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr14_+_22748980 | 0.53 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chrX_+_41193407 | 0.52 |
ENST00000457138.2
ENST00000441189.2 |
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr1_-_156675368 | 0.51 |
ENST00000368222.3
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr19_+_13842559 | 0.48 |
ENST00000586600.1
|
CCDC130
|
coiled-coil domain containing 130 |
| chr19_-_14628645 | 0.48 |
ENST00000598235.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr9_-_73029540 | 0.47 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr5_+_179247759 | 0.47 |
ENST00000389805.4
ENST00000504627.1 ENST00000402874.3 ENST00000510187.1 |
SQSTM1
|
sequestosome 1 |
| chr11_-_8832182 | 0.47 |
ENST00000527510.1
ENST00000528527.1 ENST00000528523.1 ENST00000313726.6 |
ST5
|
suppression of tumorigenicity 5 |
| chr20_+_48429356 | 0.47 |
ENST00000361573.2
ENST00000541138.1 ENST00000539601.1 |
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr18_+_61637159 | 0.46 |
ENST00000397985.2
ENST00000353706.2 ENST00000542677.1 ENST00000397988.3 ENST00000448851.1 |
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
| chr14_+_55221541 | 0.46 |
ENST00000555192.1
|
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr7_-_98467543 | 0.46 |
ENST00000345589.4
|
TMEM130
|
transmembrane protein 130 |
| chr16_+_83986827 | 0.45 |
ENST00000393306.1
ENST00000565123.1 |
OSGIN1
|
oxidative stress induced growth inhibitor 1 |
| chr11_-_76381029 | 0.45 |
ENST00000407242.2
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr7_-_98467629 | 0.44 |
ENST00000339375.4
|
TMEM130
|
transmembrane protein 130 |
| chr2_-_219134822 | 0.43 |
ENST00000444053.1
ENST00000248450.4 |
AAMP
|
angio-associated, migratory cell protein |
| chr14_-_25103388 | 0.42 |
ENST00000526004.1
ENST00000415355.3 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr3_+_191046810 | 0.42 |
ENST00000392455.3
ENST00000392456.3 |
CCDC50
|
coiled-coil domain containing 50 |
| chr16_+_57662138 | 0.41 |
ENST00000562414.1
ENST00000561969.1 ENST00000562631.1 ENST00000563445.1 ENST00000565338.1 ENST00000567702.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr3_-_32022733 | 0.41 |
ENST00000438237.2
ENST00000396556.2 |
OSBPL10
|
oxysterol binding protein-like 10 |
| chr14_-_25103472 | 0.41 |
ENST00000216341.4
ENST00000382542.1 ENST00000382540.1 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr3_+_69134080 | 0.41 |
ENST00000273258.3
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr16_+_31404624 | 0.41 |
ENST00000389202.2
|
ITGAD
|
integrin, alpha D |
| chr7_+_134528635 | 0.41 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr19_+_49377575 | 0.41 |
ENST00000600406.1
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A |
| chr3_+_19988885 | 0.41 |
ENST00000422242.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr11_+_128563948 | 0.40 |
ENST00000534087.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr17_+_61571746 | 0.40 |
ENST00000579409.1
|
ACE
|
angiotensin I converting enzyme |
| chr5_+_177540444 | 0.40 |
ENST00000274605.5
|
N4BP3
|
NEDD4 binding protein 3 |
| chr12_+_9144626 | 0.39 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr17_-_7493390 | 0.39 |
ENST00000538513.2
ENST00000570788.1 ENST00000250055.2 |
SOX15
|
SRY (sex determining region Y)-box 15 |
| chr3_+_184038073 | 0.39 |
ENST00000428387.1
ENST00000434061.2 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr1_-_244006528 | 0.39 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr3_-_187454281 | 0.38 |
ENST00000232014.4
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr4_-_102268628 | 0.38 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr19_+_36630855 | 0.38 |
ENST00000589146.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr10_+_86088337 | 0.38 |
ENST00000359979.4
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr1_+_36554470 | 0.37 |
ENST00000373178.4
|
ADPRHL2
|
ADP-ribosylhydrolase like 2 |
| chr20_+_48429233 | 0.37 |
ENST00000417961.1
|
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr11_+_128563652 | 0.37 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr7_-_98467489 | 0.37 |
ENST00000416379.2
|
TMEM130
|
transmembrane protein 130 |
| chr11_-_118134997 | 0.37 |
ENST00000278937.2
|
MPZL2
|
myelin protein zero-like 2 |
| chr5_-_138534071 | 0.36 |
ENST00000394817.2
|
SIL1
|
SIL1 nucleotide exchange factor |
| chr2_-_220173685 | 0.36 |
ENST00000423636.2
ENST00000442029.1 ENST00000412847.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr16_+_57662419 | 0.36 |
ENST00000388812.4
ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56
|
G protein-coupled receptor 56 |
| chr4_-_102268484 | 0.36 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr3_+_30648066 | 0.36 |
ENST00000359013.4
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr6_+_74405501 | 0.35 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr1_-_202777535 | 0.35 |
ENST00000367264.2
|
KDM5B
|
lysine (K)-specific demethylase 5B |
| chr3_+_45067659 | 0.35 |
ENST00000296130.4
|
CLEC3B
|
C-type lectin domain family 3, member B |
| chr11_-_65430251 | 0.35 |
ENST00000534283.1
ENST00000527749.1 ENST00000533187.1 ENST00000525693.1 ENST00000534558.1 ENST00000532879.1 ENST00000532999.1 |
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr6_+_36410762 | 0.34 |
ENST00000483557.1
ENST00000498267.1 ENST00000544295.1 ENST00000449081.2 ENST00000536244.1 ENST00000460983.1 |
KCTD20
|
potassium channel tetramerization domain containing 20 |
| chr6_+_2988847 | 0.34 |
ENST00000380472.3
ENST00000605901.1 ENST00000454015.1 |
NQO2
LINC01011
|
NAD(P)H dehydrogenase, quinone 2 long intergenic non-protein coding RNA 1011 |
| chr2_+_87755054 | 0.34 |
ENST00000423846.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr20_+_33759854 | 0.34 |
ENST00000216968.4
|
PROCR
|
protein C receptor, endothelial |
| chr20_-_634000 | 0.34 |
ENST00000381962.3
|
SRXN1
|
sulfiredoxin 1 |
| chr5_+_138089100 | 0.34 |
ENST00000520339.1
ENST00000355078.5 ENST00000302763.7 ENST00000518910.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr1_+_201159914 | 0.33 |
ENST00000335211.4
ENST00000451870.2 ENST00000295591.8 |
IGFN1
|
immunoglobulin-like and fibronectin type III domain containing 1 |
| chr14_+_103801140 | 0.33 |
ENST00000561325.1
ENST00000392715.2 ENST00000559130.1 ENST00000559532.1 ENST00000558506.1 |
EIF5
|
eukaryotic translation initiation factor 5 |
| chr6_+_74405804 | 0.33 |
ENST00000287097.5
|
CD109
|
CD109 molecule |
| chr22_+_22550113 | 0.32 |
ENST00000390285.3
|
IGLV6-57
|
immunoglobulin lambda variable 6-57 |
| chr3_+_49027308 | 0.32 |
ENST00000383729.4
ENST00000343546.4 |
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr11_-_6341844 | 0.32 |
ENST00000303927.3
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr1_+_222988406 | 0.32 |
ENST00000448808.1
ENST00000457636.1 ENST00000439440.1 |
RP11-452F19.3
|
RP11-452F19.3 |
| chr18_+_52495426 | 0.32 |
ENST00000262094.5
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr5_+_140729649 | 0.32 |
ENST00000523390.1
|
PCDHGB1
|
protocadherin gamma subfamily B, 1 |
| chr9_+_92219919 | 0.31 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr3_+_30647994 | 0.31 |
ENST00000295754.5
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr19_-_6670128 | 0.31 |
ENST00000245912.3
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr19_-_4559814 | 0.31 |
ENST00000586582.1
|
SEMA6B
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr17_+_30771279 | 0.31 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr2_+_87754887 | 0.31 |
ENST00000409054.1
ENST00000331944.6 ENST00000409139.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr5_-_138533973 | 0.31 |
ENST00000507002.1
ENST00000505830.1 ENST00000508639.1 ENST00000265195.5 |
SIL1
|
SIL1 nucleotide exchange factor |
| chr11_-_133715394 | 0.31 |
ENST00000299140.3
ENST00000532889.1 |
SPATA19
|
spermatogenesis associated 19 |
| chr8_-_38325219 | 0.31 |
ENST00000533668.1
ENST00000413133.2 ENST00000397108.4 ENST00000526742.1 ENST00000525001.1 ENST00000425967.3 ENST00000529552.1 ENST00000397113.2 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr5_+_140762268 | 0.31 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr2_+_201980961 | 0.31 |
ENST00000342795.5
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr2_+_219135115 | 0.30 |
ENST00000248451.3
ENST00000273077.4 |
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr19_-_51523412 | 0.30 |
ENST00000391805.1
ENST00000599077.1 |
KLK10
|
kallikrein-related peptidase 10 |
| chrX_+_135251835 | 0.30 |
ENST00000456445.1
|
FHL1
|
four and a half LIM domains 1 |
| chr1_+_89990431 | 0.30 |
ENST00000330947.2
ENST00000358200.4 |
LRRC8B
|
leucine rich repeat containing 8 family, member B |
| chr12_+_9142131 | 0.30 |
ENST00000356986.3
ENST00000266551.4 |
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr7_+_128095945 | 0.30 |
ENST00000257696.4
|
HILPDA
|
hypoxia inducible lipid droplet-associated |
| chr8_+_126442563 | 0.30 |
ENST00000311922.3
|
TRIB1
|
tribbles pseudokinase 1 |
| chr1_+_222988363 | 0.30 |
ENST00000450784.1
ENST00000426045.1 ENST00000457955.1 ENST00000444858.1 ENST00000435378.1 ENST00000441676.1 |
RP11-452F19.3
|
RP11-452F19.3 |
| chr2_+_87754989 | 0.30 |
ENST00000409898.2
ENST00000419680.2 ENST00000414584.1 ENST00000455131.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr7_+_128095900 | 0.30 |
ENST00000435296.2
|
HILPDA
|
hypoxia inducible lipid droplet-associated |
| chr7_+_112063192 | 0.30 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr1_-_160232312 | 0.29 |
ENST00000440682.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr3_+_127770455 | 0.29 |
ENST00000464451.1
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr11_+_59522837 | 0.29 |
ENST00000437946.2
|
STX3
|
syntaxin 3 |
| chr19_-_51523275 | 0.29 |
ENST00000309958.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr19_-_44174330 | 0.29 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr5_+_169931009 | 0.29 |
ENST00000328939.4
ENST00000390656.4 |
KCNIP1
|
Kv channel interacting protein 1 |
| chr10_-_118928543 | 0.29 |
ENST00000419373.2
|
RP11-501J20.2
|
RP11-501J20.2 |
| chr22_-_37584321 | 0.29 |
ENST00000397110.2
ENST00000337843.2 |
C1QTNF6
|
C1q and tumor necrosis factor related protein 6 |
| chr6_-_32191834 | 0.28 |
ENST00000375023.3
|
NOTCH4
|
notch 4 |
| chr1_-_51796987 | 0.28 |
ENST00000262676.5
|
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr17_-_27503770 | 0.28 |
ENST00000533112.1
|
MYO18A
|
myosin XVIIIA |
| chr14_+_103800513 | 0.28 |
ENST00000560338.1
ENST00000560763.1 ENST00000558316.1 ENST00000558265.1 |
EIF5
|
eukaryotic translation initiation factor 5 |
| chr20_+_30946106 | 0.27 |
ENST00000375687.4
ENST00000542461.1 |
ASXL1
|
additional sex combs like 1 (Drosophila) |
| chr20_+_4667094 | 0.27 |
ENST00000424424.1
ENST00000457586.1 |
PRNP
|
prion protein |
| chr17_+_74381343 | 0.27 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr7_+_150688083 | 0.27 |
ENST00000297494.3
|
NOS3
|
nitric oxide synthase 3 (endothelial cell) |
| chr8_-_59572093 | 0.27 |
ENST00000427130.2
|
NSMAF
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr11_+_57308979 | 0.27 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr11_-_62323702 | 0.27 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr1_-_21059029 | 0.27 |
ENST00000444387.2
ENST00000375031.1 ENST00000518294.1 |
SH2D5
|
SH2 domain containing 5 |
| chr12_+_10365404 | 0.27 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr2_-_175712270 | 0.27 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr1_+_222988464 | 0.27 |
ENST00000420335.1
|
RP11-452F19.3
|
RP11-452F19.3 |
| chr17_+_35851570 | 0.26 |
ENST00000394386.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr19_+_41949054 | 0.26 |
ENST00000378187.2
|
C19orf69
|
chromosome 19 open reading frame 69 |
| chrX_+_135251783 | 0.26 |
ENST00000394153.2
|
FHL1
|
four and a half LIM domains 1 |
| chr6_-_35888824 | 0.26 |
ENST00000361690.3
ENST00000512445.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr10_-_21463116 | 0.26 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chrX_-_65253506 | 0.25 |
ENST00000427538.1
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr8_-_145025044 | 0.25 |
ENST00000322810.4
|
PLEC
|
plectin |
| chr8_-_27472198 | 0.25 |
ENST00000519472.1
ENST00000523589.1 ENST00000522413.1 ENST00000523396.1 ENST00000560366.1 |
CLU
|
clusterin |
| chr4_+_76481258 | 0.25 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr2_+_89890533 | 0.25 |
ENST00000429992.2
|
IGKV2D-40
|
immunoglobulin kappa variable 2D-40 |
| chr3_-_98241358 | 0.25 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr6_+_151042224 | 0.25 |
ENST00000358517.2
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr6_-_35888905 | 0.25 |
ENST00000510290.1
ENST00000423325.2 ENST00000373822.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr3_-_142720267 | 0.25 |
ENST00000597953.1
|
RP11-91G21.1
|
RP11-91G21.1 |
| chr7_-_130598059 | 0.24 |
ENST00000432045.2
|
MIR29B1
|
microRNA 29a |
| chr17_-_73781567 | 0.24 |
ENST00000586607.1
|
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr19_-_44008863 | 0.24 |
ENST00000601646.1
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3 |
| chr11_+_66742742 | 0.24 |
ENST00000308963.4
|
C11orf86
|
chromosome 11 open reading frame 86 |
| chrX_-_150067069 | 0.24 |
ENST00000466436.1
|
CD99L2
|
CD99 molecule-like 2 |
| chr1_-_111148241 | 0.24 |
ENST00000440270.1
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr2_-_89340242 | 0.24 |
ENST00000480492.1
|
IGKV1-12
|
immunoglobulin kappa variable 1-12 |
| chr3_+_5020801 | 0.24 |
ENST00000256495.3
|
BHLHE40
|
basic helix-loop-helix family, member e40 |
| chr8_+_104384616 | 0.23 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr11_+_66360665 | 0.23 |
ENST00000310190.4
|
CCS
|
copper chaperone for superoxide dismutase |
| chr3_+_50284321 | 0.23 |
ENST00000451956.1
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr7_+_55177416 | 0.23 |
ENST00000450046.1
ENST00000454757.2 |
EGFR
|
epidermal growth factor receptor |
| chr12_+_56862301 | 0.23 |
ENST00000338146.5
|
SPRYD4
|
SPRY domain containing 4 |
| chr12_-_39836772 | 0.23 |
ENST00000541463.2
ENST00000361418.5 ENST00000544797.2 |
KIF21A
|
kinesin family member 21A |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOSL2_SMARCC1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 1.0 | 2.9 | GO:0072019 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.5 | 1.8 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.4 | 1.5 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.4 | 1.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 | 0.9 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.3 | 2.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.2 | 2.7 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 0.7 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.2 | 2.3 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.2 | 0.7 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.2 | 0.6 | GO:0034146 | B-1 B cell homeostasis(GO:0001922) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) negative regulation of osteoclast proliferation(GO:0090291) |
| 0.2 | 0.6 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.2 | 0.4 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.2 | 1.9 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.2 | 1.1 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.2 | 1.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.2 | 0.7 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.2 | 2.6 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.6 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 1.0 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.4 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) smooth muscle hyperplasia(GO:0014806) |
| 0.1 | 0.4 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 0.7 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) terminal web assembly(GO:1902896) |
| 0.1 | 0.4 | GO:0048627 | myoblast development(GO:0048627) |
| 0.1 | 0.8 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 0.4 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 0.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.1 | 0.7 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.6 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.1 | 0.8 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.1 | 1.0 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.4 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 0.4 | GO:2000864 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.1 | 0.3 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.1 | 0.3 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.4 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.1 | 1.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.4 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.1 | 0.3 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.7 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 0.3 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 0.4 | GO:0033123 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.1 | 0.3 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.3 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.3 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 0.4 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.1 | 0.3 | GO:0014040 | positive regulation of Schwann cell differentiation(GO:0014040) |
| 0.1 | 0.5 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.5 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.1 | 0.4 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.1 | 0.2 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.1 | 0.4 | GO:0001554 | luteolysis(GO:0001554) |
| 0.1 | 0.2 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.1 | 0.5 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.1 | 1.0 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 0.2 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 0.2 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 0.6 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.2 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.1 | 0.3 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) neuron projection maintenance(GO:1990535) |
| 0.1 | 0.7 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.1 | 0.5 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.2 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.1 | 0.5 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 0.3 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.3 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.2 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.1 | 0.2 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 0.2 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.1 | 0.2 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.1 | 0.2 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 0.2 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.2 | GO:0060545 | CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) positive regulation of necroptotic process(GO:0060545) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.1 | 0.2 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.1 | GO:0032759 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.0 | 0.3 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.2 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 0.8 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.0 | 0.6 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.7 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.3 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.2 | GO:1902996 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) regulation of neurofibrillary tangle assembly(GO:1902996) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.6 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.2 | GO:0033484 | nitric oxide homeostasis(GO:0033484) response to L-ascorbic acid(GO:0033591) negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.5 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.2 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 1.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.3 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.5 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.2 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.2 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:1901094 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.0 | 0.1 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.7 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.0 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.1 | GO:0001983 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) positive regulation of the force of heart contraction by chemical signal(GO:0003099) pilomotor reflex(GO:0097195) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.3 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.9 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.2 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0086098 | angiotensin-activated signaling pathway involved in heart process(GO:0086098) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.5 | GO:0002251 | organ or tissue specific immune response(GO:0002251) mucosal immune response(GO:0002385) |
| 0.0 | 0.1 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:1904995 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.1 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.1 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.3 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.4 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.0 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) |
| 0.0 | 0.2 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.0 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.0 | 0.5 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.2 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.5 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.2 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.4 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.1 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.2 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.3 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.2 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.7 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 0.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.5 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.3 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 0.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.0 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.0 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.0 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.5 | GO:0071451 | cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.0 | 0.8 | GO:0006968 | cellular defense response(GO:0006968) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.7 | 5.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.5 | 1.8 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.2 | 0.7 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.2 | 0.7 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.2 | 0.5 | GO:0044753 | amphisome(GO:0044753) |
| 0.2 | 0.6 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 0.5 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 0.3 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.1 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.9 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 0.7 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.2 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.1 | 0.7 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.3 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 1.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.2 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.1 | 0.4 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 1.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 1.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 1.0 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) outer acrosomal membrane(GO:0002081) |
| 0.0 | 4.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 2.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.4 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 1.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 2.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 1.0 | 2.9 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.6 | 1.8 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.3 | 1.5 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.2 | 2.7 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.2 | 0.9 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 1.0 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 0.6 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.2 | 0.7 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.2 | 1.7 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.4 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.1 | 0.4 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.6 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.3 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 1.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.6 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.1 | 0.3 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.1 | 0.2 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.1 | 0.3 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.7 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.3 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.1 | 0.8 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.4 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.2 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 0.2 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.1 | 0.4 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.2 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.1 | 0.2 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.1 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.2 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.4 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 1.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.2 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.5 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.0 | 0.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 1.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.4 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.2 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.4 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.7 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.3 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 4.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.0 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.1 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 3.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 4.9 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.7 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 3.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.9 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.0 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 5.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 0.2 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 3.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 1.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 2.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 1.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.6 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.8 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.4 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.6 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.5 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 1.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.0 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |