Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for FOXF2_FOXJ1
Z-value: 0.42
Transcription factors associated with FOXF2_FOXJ1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
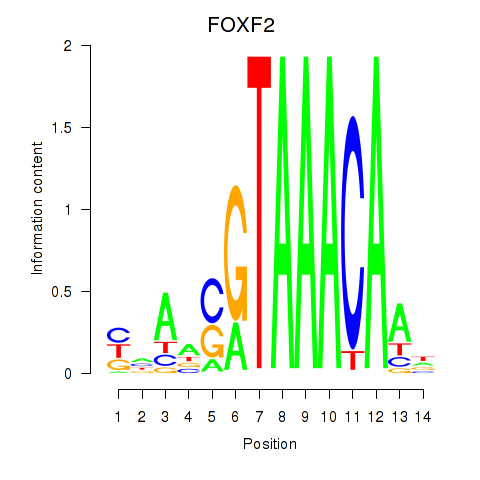
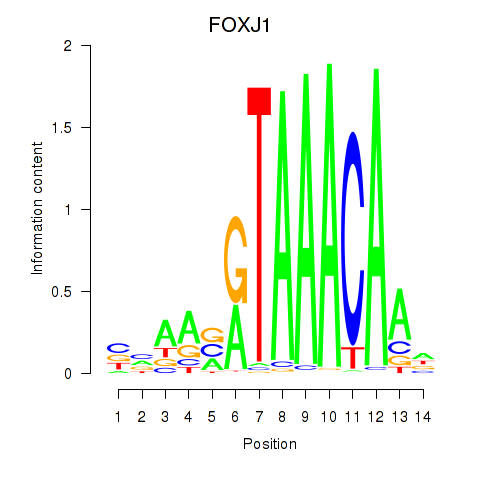
FOXF2
|
ENSG00000137273.3 | forkhead box F2 |
|
FOXJ1
|
ENSG00000129654.7 | forkhead box J1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXJ1 | hg19_v2_chr17_-_74137374_74137385 | -0.10 | 6.5e-01 | Click! |
| FOXF2 | hg19_v2_chr6_+_1389989_1390069 | 0.02 | 9.1e-01 | Click! |
Activity profile of FOXF2_FOXJ1 motif
Sorted Z-values of FOXF2_FOXJ1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_129906660 | 0.77 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr15_-_40600026 | 0.55 |
ENST00000456256.2
ENST00000557821.1 |
PLCB2
|
phospholipase C, beta 2 |
| chr2_-_157198860 | 0.48 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr8_+_11666649 | 0.46 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr4_+_100495864 | 0.38 |
ENST00000265517.5
ENST00000422897.2 |
MTTP
|
microsomal triglyceride transfer protein |
| chr1_+_185703513 | 0.37 |
ENST00000271588.4
ENST00000367492.2 |
HMCN1
|
hemicentin 1 |
| chr13_-_99910673 | 0.34 |
ENST00000397473.2
ENST00000397470.2 |
GPR18
|
G protein-coupled receptor 18 |
| chr7_-_122635754 | 0.34 |
ENST00000249284.2
|
TAS2R16
|
taste receptor, type 2, member 16 |
| chr8_-_134309335 | 0.32 |
ENST00000522890.1
ENST00000323851.7 ENST00000518176.1 ENST00000354944.5 ENST00000537882.1 ENST00000522476.1 ENST00000518066.1 ENST00000521544.1 ENST00000518480.1 ENST00000523892.1 |
NDRG1
|
N-myc downstream regulated 1 |
| chr8_-_134309823 | 0.32 |
ENST00000414097.2
|
NDRG1
|
N-myc downstream regulated 1 |
| chr2_+_33661382 | 0.31 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr19_+_41768561 | 0.28 |
ENST00000599719.1
ENST00000601309.1 |
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr16_+_77233294 | 0.27 |
ENST00000378644.4
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr8_-_124553437 | 0.27 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr8_+_67344710 | 0.26 |
ENST00000379385.4
ENST00000396623.3 ENST00000415254.1 |
ADHFE1
|
alcohol dehydrogenase, iron containing, 1 |
| chr16_-_86542652 | 0.24 |
ENST00000599749.1
|
FENDRR
|
FOXF1 adjacent non-coding developmental regulatory RNA |
| chr15_-_31283798 | 0.24 |
ENST00000435680.1
ENST00000425768.1 |
MTMR10
|
myotubularin related protein 10 |
| chr15_+_84906338 | 0.24 |
ENST00000512109.1
|
GOLGA6L4
|
golgin A6 family-like 4 |
| chr19_+_50380682 | 0.23 |
ENST00000221543.5
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr20_+_44035847 | 0.23 |
ENST00000372712.2
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr19_+_50380917 | 0.23 |
ENST00000535102.2
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr4_-_111119804 | 0.23 |
ENST00000394607.3
ENST00000302274.3 |
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr4_-_123542224 | 0.22 |
ENST00000264497.3
|
IL21
|
interleukin 21 |
| chr20_+_44035200 | 0.22 |
ENST00000372717.1
ENST00000360981.4 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr5_+_60933634 | 0.22 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr4_-_70626314 | 0.20 |
ENST00000510821.1
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr10_-_14050522 | 0.20 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr12_-_102591604 | 0.20 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr20_+_45338126 | 0.19 |
ENST00000359271.2
|
SLC2A10
|
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr4_+_146403912 | 0.19 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr2_+_12857043 | 0.19 |
ENST00000381465.2
|
TRIB2
|
tribbles pseudokinase 2 |
| chr9_-_135819987 | 0.19 |
ENST00000298552.3
ENST00000403810.1 |
TSC1
|
tuberous sclerosis 1 |
| chr15_+_28624878 | 0.18 |
ENST00000450328.2
|
GOLGA8F
|
golgin A8 family, member F |
| chr6_+_72922590 | 0.18 |
ENST00000523963.1
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr6_+_72922505 | 0.18 |
ENST00000401910.3
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr7_-_105332084 | 0.17 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr6_-_76203345 | 0.17 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr6_+_21666633 | 0.17 |
ENST00000606851.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr12_-_63328817 | 0.16 |
ENST00000228705.6
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr19_-_36523709 | 0.16 |
ENST00000592017.1
ENST00000360535.4 |
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr2_-_220042825 | 0.16 |
ENST00000409789.1
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr5_-_42812143 | 0.16 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr1_+_152486950 | 0.16 |
ENST00000368790.3
|
CRCT1
|
cysteine-rich C-terminal 1 |
| chr2_+_233527443 | 0.16 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr12_-_15038779 | 0.15 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr7_-_25702669 | 0.15 |
ENST00000446840.1
|
AC003090.1
|
AC003090.1 |
| chr9_+_139702363 | 0.15 |
ENST00000371663.4
ENST00000371671.4 ENST00000311502.7 |
RABL6
|
RAB, member RAS oncogene family-like 6 |
| chr11_+_108535849 | 0.15 |
ENST00000526794.1
|
DDX10
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
| chr9_-_5185629 | 0.15 |
ENST00000381641.3
|
INSL6
|
insulin-like 6 |
| chr8_+_120079478 | 0.14 |
ENST00000332843.2
|
COLEC10
|
collectin sub-family member 10 (C-type lectin) |
| chr7_+_134832808 | 0.14 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr6_-_2751146 | 0.14 |
ENST00000268446.5
ENST00000274643.7 |
MYLK4
|
myosin light chain kinase family, member 4 |
| chr21_+_17443434 | 0.14 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_+_22351977 | 0.14 |
ENST00000420503.1
ENST00000416769.1 ENST00000404210.2 |
LINC00339
|
long intergenic non-protein coding RNA 339 |
| chr12_-_56352368 | 0.13 |
ENST00000549404.1
|
PMEL
|
premelanosome protein |
| chr8_+_97597148 | 0.13 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr5_+_81601166 | 0.13 |
ENST00000439350.1
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr20_-_62582475 | 0.13 |
ENST00000369908.5
|
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr7_+_5229819 | 0.13 |
ENST00000288828.4
ENST00000401525.3 ENST00000404704.3 |
WIPI2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr1_-_57431679 | 0.13 |
ENST00000371237.4
ENST00000535057.1 ENST00000543257.1 |
C8B
|
complement component 8, beta polypeptide |
| chr12_-_92539614 | 0.13 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr15_+_75940218 | 0.12 |
ENST00000308527.5
|
SNX33
|
sorting nexin 33 |
| chr5_-_180076580 | 0.12 |
ENST00000502649.1
|
FLT4
|
fms-related tyrosine kinase 4 |
| chr17_-_39150385 | 0.12 |
ENST00000391586.1
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr11_-_114466471 | 0.12 |
ENST00000424261.2
|
NXPE4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr8_-_6420930 | 0.12 |
ENST00000325203.5
|
ANGPT2
|
angiopoietin 2 |
| chr11_-_34535297 | 0.12 |
ENST00000532417.1
|
ELF5
|
E74-like factor 5 (ets domain transcription factor) |
| chr8_-_6420777 | 0.12 |
ENST00000415216.1
|
ANGPT2
|
angiopoietin 2 |
| chr5_-_180076613 | 0.12 |
ENST00000261937.6
ENST00000393347.3 |
FLT4
|
fms-related tyrosine kinase 4 |
| chr3_+_136676851 | 0.12 |
ENST00000309741.5
|
IL20RB
|
interleukin 20 receptor beta |
| chr6_+_43027595 | 0.12 |
ENST00000259708.3
ENST00000472792.1 ENST00000479388.1 ENST00000460283.1 ENST00000394056.2 |
KLC4
|
kinesin light chain 4 |
| chr4_-_70626430 | 0.12 |
ENST00000310613.3
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr6_-_42016385 | 0.12 |
ENST00000502771.1
ENST00000508143.1 ENST00000514588.1 ENST00000510503.1 ENST00000415497.2 ENST00000372988.4 |
CCND3
|
cyclin D3 |
| chr7_+_5229904 | 0.12 |
ENST00000382384.2
|
WIPI2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr1_+_47533160 | 0.11 |
ENST00000334194.3
|
CYP4Z1
|
cytochrome P450, family 4, subfamily Z, polypeptide 1 |
| chr14_-_36988882 | 0.11 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr3_+_136676707 | 0.11 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr18_-_53068911 | 0.11 |
ENST00000537856.3
|
TCF4
|
transcription factor 4 |
| chr14_+_50234827 | 0.11 |
ENST00000554589.1
ENST00000557247.1 |
KLHDC2
|
kelch domain containing 2 |
| chr3_-_168865522 | 0.11 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr2_+_86947296 | 0.11 |
ENST00000283632.4
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| chr10_-_73848086 | 0.11 |
ENST00000536168.1
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr7_+_44646218 | 0.11 |
ENST00000444676.1
ENST00000222673.5 |
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr16_-_28374829 | 0.11 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family, member B6 |
| chr11_-_63933504 | 0.11 |
ENST00000255681.6
|
MACROD1
|
MACRO domain containing 1 |
| chr16_+_6069586 | 0.11 |
ENST00000547372.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr17_+_58755184 | 0.10 |
ENST00000589222.1
ENST00000407086.3 ENST00000390652.5 |
BCAS3
|
breast carcinoma amplified sequence 3 |
| chr11_-_27723158 | 0.10 |
ENST00000395980.2
|
BDNF
|
brain-derived neurotrophic factor |
| chr1_+_74701062 | 0.10 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr9_-_86432547 | 0.10 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr16_+_6069072 | 0.10 |
ENST00000547605.1
ENST00000550418.1 ENST00000553186.1 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr21_+_17443521 | 0.10 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr20_+_59654146 | 0.10 |
ENST00000441660.1
|
RP5-827L5.1
|
RP5-827L5.1 |
| chr12_+_40787194 | 0.10 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr5_-_36301984 | 0.10 |
ENST00000502994.1
ENST00000515759.1 ENST00000296604.3 |
RANBP3L
|
RAN binding protein 3-like |
| chr1_+_150122034 | 0.10 |
ENST00000025469.6
ENST00000369124.4 |
PLEKHO1
|
pleckstrin homology domain containing, family O member 1 |
| chr9_-_134585221 | 0.10 |
ENST00000372190.3
ENST00000427994.1 |
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr3_+_132316081 | 0.09 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr6_-_76203454 | 0.09 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr17_-_8263538 | 0.09 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr2_-_220264703 | 0.09 |
ENST00000519905.1
ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr4_+_71108300 | 0.09 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr10_-_13570533 | 0.09 |
ENST00000396900.2
ENST00000396898.2 |
BEND7
|
BEN domain containing 7 |
| chr11_+_34654011 | 0.09 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr16_+_28763108 | 0.08 |
ENST00000357796.3
ENST00000550983.1 |
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr11_-_34535332 | 0.08 |
ENST00000257832.2
ENST00000429939.2 |
ELF5
|
E74-like factor 5 (ets domain transcription factor) |
| chr8_-_145754428 | 0.08 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.1 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chrX_+_85969626 | 0.08 |
ENST00000484479.1
|
DACH2
|
dachshund homolog 2 (Drosophila) |
| chr3_+_137717571 | 0.08 |
ENST00000343735.4
|
CLDN18
|
claudin 18 |
| chr6_+_101846664 | 0.08 |
ENST00000421544.1
ENST00000413795.1 ENST00000369138.1 ENST00000358361.3 |
GRIK2
|
glutamate receptor, ionotropic, kainate 2 |
| chr2_+_220042933 | 0.08 |
ENST00000430297.2
|
FAM134A
|
family with sequence similarity 134, member A |
| chr6_+_43027332 | 0.08 |
ENST00000347162.5
ENST00000453940.2 ENST00000479632.1 ENST00000470728.1 ENST00000458460.2 |
KLC4
|
kinesin light chain 4 |
| chr2_+_179184955 | 0.08 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr6_+_84563295 | 0.08 |
ENST00000369687.1
|
RIPPLY2
|
ripply transcriptional repressor 2 |
| chr1_+_87797351 | 0.08 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr14_-_101036119 | 0.08 |
ENST00000355173.2
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr11_+_120973375 | 0.07 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr17_+_72427477 | 0.07 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chrX_-_47863348 | 0.07 |
ENST00000376943.3
ENST00000396965.1 ENST00000305127.6 |
ZNF182
|
zinc finger protein 182 |
| chr6_+_32121789 | 0.07 |
ENST00000437001.2
ENST00000375137.2 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr5_-_19988339 | 0.07 |
ENST00000382275.1
|
CDH18
|
cadherin 18, type 2 |
| chr20_-_44993012 | 0.07 |
ENST00000372229.1
ENST00000372230.5 ENST00000543605.1 ENST00000243896.2 ENST00000317734.8 |
SLC35C2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr6_+_32121908 | 0.07 |
ENST00000375143.2
ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr17_+_72426891 | 0.07 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr20_+_33563206 | 0.07 |
ENST00000262873.7
|
MYH7B
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr22_-_39268308 | 0.07 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr9_-_77502636 | 0.07 |
ENST00000449912.2
|
TRPM6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr16_-_80926457 | 0.07 |
ENST00000563626.1
ENST00000562231.1 |
RP11-314O13.1
|
RP11-314O13.1 |
| chr11_-_85397167 | 0.07 |
ENST00000316398.3
|
CCDC89
|
coiled-coil domain containing 89 |
| chr2_+_58655461 | 0.07 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr2_-_160472952 | 0.07 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr4_-_21950356 | 0.06 |
ENST00000447367.2
ENST00000382152.2 |
KCNIP4
|
Kv channel interacting protein 4 |
| chr20_-_25320367 | 0.06 |
ENST00000450393.1
ENST00000491682.1 |
ABHD12
|
abhydrolase domain containing 12 |
| chr17_-_40134339 | 0.06 |
ENST00000587727.1
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr19_-_50380536 | 0.06 |
ENST00000391832.3
ENST00000391834.2 ENST00000344175.5 |
AKT1S1
|
AKT1 substrate 1 (proline-rich) |
| chrX_-_65259900 | 0.06 |
ENST00000412866.2
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr4_-_70518941 | 0.06 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr21_+_17442799 | 0.06 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr11_-_118972575 | 0.06 |
ENST00000432443.2
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr10_-_118032697 | 0.06 |
ENST00000439649.3
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr2_-_233877912 | 0.06 |
ENST00000264051.3
|
NGEF
|
neuronal guanine nucleotide exchange factor |
| chrX_-_65259914 | 0.06 |
ENST00000374737.4
ENST00000455586.2 |
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chrX_+_135388147 | 0.06 |
ENST00000394141.1
|
GPR112
|
G protein-coupled receptor 112 |
| chr19_-_47157914 | 0.06 |
ENST00000300875.4
|
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr3_-_10547192 | 0.06 |
ENST00000360273.2
ENST00000343816.4 |
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr5_+_176853702 | 0.06 |
ENST00000507633.1
ENST00000393576.3 ENST00000355958.5 ENST00000528793.1 ENST00000512684.1 |
GRK6
|
G protein-coupled receptor kinase 6 |
| chr3_+_159570722 | 0.06 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr11_+_844406 | 0.06 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr4_-_123377880 | 0.06 |
ENST00000226730.4
|
IL2
|
interleukin 2 |
| chr5_+_140529630 | 0.06 |
ENST00000543635.1
|
PCDHB6
|
protocadherin beta 6 |
| chr4_-_90757364 | 0.06 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr18_+_18943554 | 0.06 |
ENST00000580732.2
|
GREB1L
|
growth regulation by estrogen in breast cancer-like |
| chr6_-_112575838 | 0.05 |
ENST00000455073.1
|
LAMA4
|
laminin, alpha 4 |
| chr5_-_110062384 | 0.05 |
ENST00000429839.2
|
TMEM232
|
transmembrane protein 232 |
| chr19_-_10446449 | 0.05 |
ENST00000592439.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr11_-_115375107 | 0.05 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr3_-_10547333 | 0.05 |
ENST00000383800.4
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr14_+_23299088 | 0.05 |
ENST00000355151.5
ENST00000397496.3 ENST00000555345.1 ENST00000432849.3 ENST00000553711.1 ENST00000556465.1 ENST00000397505.2 ENST00000557221.1 ENST00000311892.6 ENST00000556840.1 ENST00000555536.1 |
MRPL52
|
mitochondrial ribosomal protein L52 |
| chr5_-_110062349 | 0.05 |
ENST00000511883.2
ENST00000455884.2 |
TMEM232
|
transmembrane protein 232 |
| chr2_+_162272605 | 0.05 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr2_-_14541060 | 0.05 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr12_-_75603202 | 0.05 |
ENST00000393288.2
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr5_+_154092396 | 0.05 |
ENST00000336314.4
|
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr20_-_22566089 | 0.05 |
ENST00000377115.4
|
FOXA2
|
forkhead box A2 |
| chr3_+_119298523 | 0.05 |
ENST00000357003.3
|
ADPRH
|
ADP-ribosylarginine hydrolase |
| chr4_-_90756769 | 0.05 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr19_+_13906250 | 0.05 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr22_-_39928823 | 0.05 |
ENST00000334678.3
|
RPS19BP1
|
ribosomal protein S19 binding protein 1 |
| chr5_+_176853669 | 0.04 |
ENST00000355472.5
|
GRK6
|
G protein-coupled receptor kinase 6 |
| chr12_-_96390063 | 0.04 |
ENST00000541929.1
|
HAL
|
histidine ammonia-lyase |
| chr2_-_180610767 | 0.04 |
ENST00000409343.1
|
ZNF385B
|
zinc finger protein 385B |
| chr4_+_71588372 | 0.04 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr5_+_140734570 | 0.04 |
ENST00000571252.1
|
PCDHGA4
|
protocadherin gamma subfamily A, 4 |
| chr3_-_58523010 | 0.04 |
ENST00000459701.2
ENST00000302819.5 |
ACOX2
|
acyl-CoA oxidase 2, branched chain |
| chr16_-_84178728 | 0.04 |
ENST00000562224.1
ENST00000434463.3 ENST00000564998.1 ENST00000219439.4 |
HSDL1
|
hydroxysteroid dehydrogenase like 1 |
| chr3_-_48956818 | 0.04 |
ENST00000408959.2
|
ARIH2OS
|
ariadne homolog 2 opposite strand |
| chr3_+_98451532 | 0.04 |
ENST00000486334.2
ENST00000394162.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr11_+_844067 | 0.04 |
ENST00000397406.1
ENST00000409543.2 ENST00000525201.1 |
TSPAN4
|
tetraspanin 4 |
| chr4_+_71587669 | 0.04 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr9_+_78505581 | 0.04 |
ENST00000376767.3
ENST00000376752.4 |
PCSK5
|
proprotein convertase subtilisin/kexin type 5 |
| chr1_+_246729724 | 0.04 |
ENST00000366513.4
ENST00000366512.3 |
CNST
|
consortin, connexin sorting protein |
| chr11_+_27076764 | 0.04 |
ENST00000525090.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr5_-_42811986 | 0.04 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr1_-_149982624 | 0.04 |
ENST00000417191.1
ENST00000369135.4 |
OTUD7B
|
OTU domain containing 7B |
| chr16_+_84178874 | 0.04 |
ENST00000378553.5
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr1_+_81771806 | 0.04 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr14_+_28081790 | 0.04 |
ENST00000557359.1
ENST00000557399.1 |
LINC00645
|
long intergenic non-protein coding RNA 645 |
| chr2_-_175260368 | 0.04 |
ENST00000342016.3
ENST00000362053.5 |
CIR1
|
corepressor interacting with RBPJ, 1 |
| chr3_+_48956249 | 0.04 |
ENST00000452882.1
ENST00000430423.1 ENST00000356401.4 ENST00000449376.1 ENST00000420814.1 ENST00000449729.1 ENST00000433170.1 |
ARIH2
|
ariadne RBR E3 ubiquitin protein ligase 2 |
| chr7_+_151038850 | 0.04 |
ENST00000355851.4
ENST00000566856.1 ENST00000470229.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr1_+_92545862 | 0.04 |
ENST00000370382.3
ENST00000342818.3 |
BTBD8
|
BTB (POZ) domain containing 8 |
| chr17_+_58677539 | 0.04 |
ENST00000305921.3
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D |
| chr12_-_49449107 | 0.04 |
ENST00000301067.7
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr6_+_161123270 | 0.04 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr17_+_16284604 | 0.04 |
ENST00000395839.1
ENST00000395837.1 |
UBB
|
ubiquitin B |
| chr7_-_140340576 | 0.04 |
ENST00000275884.6
ENST00000475837.1 |
DENND2A
|
DENN/MADD domain containing 2A |
| chr17_+_67410832 | 0.03 |
ENST00000590474.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr1_-_246729544 | 0.03 |
ENST00000544618.1
ENST00000366514.4 |
TFB2M
|
transcription factor B2, mitochondrial |
| chr19_-_49016418 | 0.03 |
ENST00000270238.3
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr4_-_140223614 | 0.03 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXF2_FOXJ1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 0.5 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 0.3 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.1 | 0.3 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.1 | 0.6 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 0.2 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.2 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.2 | GO:0051029 | rRNA transport(GO:0051029) |
| 0.0 | 0.8 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.2 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.4 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.3 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.2 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.3 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0051622 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.4 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.2 | GO:0060312 | regulation of blood vessel remodeling(GO:0060312) |
| 0.0 | 0.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.0 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.7 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.0 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.0 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.0 | 0.0 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 | 0.1 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.3 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.0 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.3 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.0 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.2 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.3 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.0 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.0 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.0 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.0 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.0 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |