Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
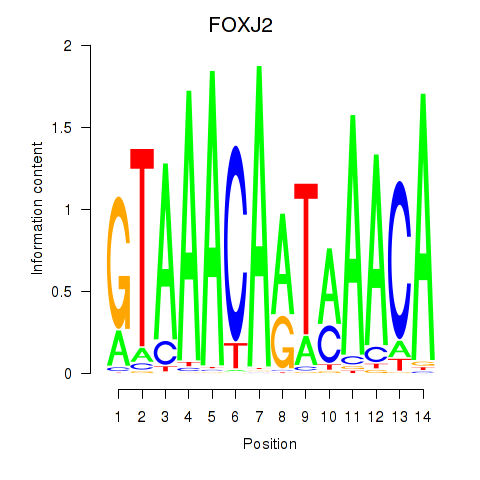
Results for FOXJ2
Z-value: 0.81
Transcription factors associated with FOXJ2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXJ2
|
ENSG00000065970.4 | forkhead box J2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXJ2 | hg19_v2_chr12_+_8185288_8185339 | -0.37 | 6.6e-02 | Click! |
Activity profile of FOXJ2 motif
Sorted Z-values of FOXJ2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_67430339 | 4.78 |
ENST00000439724.3
|
SMAD3
|
SMAD family member 3 |
| chr8_+_104383728 | 2.19 |
ENST00000330295.5
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr2_+_162016827 | 2.02 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr11_+_35201826 | 1.74 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr1_+_165797024 | 1.71 |
ENST00000372212.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr9_+_5510492 | 1.59 |
ENST00000397745.2
|
PDCD1LG2
|
programmed cell death 1 ligand 2 |
| chr9_+_5510558 | 1.55 |
ENST00000397747.3
|
PDCD1LG2
|
programmed cell death 1 ligand 2 |
| chr2_+_162016804 | 1.41 |
ENST00000392749.2
ENST00000440506.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr21_-_35899113 | 1.41 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr9_-_134615443 | 1.13 |
ENST00000372195.1
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr7_-_115608304 | 1.11 |
ENST00000457268.1
|
TFEC
|
transcription factor EC |
| chr2_+_162016916 | 1.10 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr17_+_25958174 | 1.07 |
ENST00000313648.6
ENST00000577392.1 ENST00000584661.1 ENST00000413914.2 |
LGALS9
|
lectin, galactoside-binding, soluble, 9 |
| chr7_-_80548667 | 1.04 |
ENST00000265361.3
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr10_+_28822417 | 0.81 |
ENST00000428935.1
ENST00000420266.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr21_-_27423339 | 0.81 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr1_-_43855444 | 0.77 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr10_+_28822636 | 0.75 |
ENST00000442148.1
ENST00000448193.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr10_-_36813162 | 0.74 |
ENST00000440465.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr11_-_57089671 | 0.71 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chrX_+_108780347 | 0.70 |
ENST00000372103.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr19_+_7030589 | 0.69 |
ENST00000329753.5
|
MBD3L5
|
methyl-CpG binding domain protein 3-like 5 |
| chr10_+_33271469 | 0.67 |
ENST00000414157.1
|
RP11-462L8.1
|
RP11-462L8.1 |
| chr6_+_30539153 | 0.66 |
ENST00000326195.8
ENST00000376545.3 ENST00000396515.4 ENST00000441867.1 ENST00000468958.1 |
ABCF1
|
ATP-binding cassette, sub-family F (GCN20), member 1 |
| chr1_+_150122034 | 0.66 |
ENST00000025469.6
ENST00000369124.4 |
PLEKHO1
|
pleckstrin homology domain containing, family O member 1 |
| chr10_+_118305435 | 0.65 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr2_-_239140011 | 0.61 |
ENST00000409376.1
ENST00000409070.1 ENST00000409942.1 |
AC016757.3
|
Protein LOC151174 |
| chr19_-_55677920 | 0.60 |
ENST00000524407.2
ENST00000526003.1 ENST00000534170.1 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr3_+_171757346 | 0.59 |
ENST00000421757.1
ENST00000415807.2 ENST00000392699.1 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr12_+_54891495 | 0.59 |
ENST00000293373.6
|
NCKAP1L
|
NCK-associated protein 1-like |
| chr2_-_239140276 | 0.57 |
ENST00000334973.4
|
AC016757.3
|
Protein LOC151174 |
| chr15_+_66797627 | 0.57 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chrX_-_6453159 | 0.56 |
ENST00000381089.3
ENST00000398729.1 |
VCX3A
|
variable charge, X-linked 3A |
| chr3_-_190167571 | 0.52 |
ENST00000354905.2
|
TMEM207
|
transmembrane protein 207 |
| chr6_+_11094266 | 0.52 |
ENST00000416247.2
|
SMIM13
|
small integral membrane protein 13 |
| chr19_-_7058651 | 0.52 |
ENST00000333843.4
|
MBD3L3
|
methyl-CpG binding domain protein 3-like 3 |
| chr17_-_39324424 | 0.52 |
ENST00000391356.2
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr12_-_10251539 | 0.51 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr20_-_42355629 | 0.50 |
ENST00000373003.1
|
GTSF1L
|
gametocyte specific factor 1-like |
| chr7_-_102283238 | 0.50 |
ENST00000340457.8
|
UPK3BL
|
uroplakin 3B-like |
| chr1_-_72566613 | 0.50 |
ENST00000306821.3
|
NEGR1
|
neuronal growth regulator 1 |
| chr4_-_47983519 | 0.49 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr21_-_37451680 | 0.49 |
ENST00000399201.1
|
SETD4
|
SET domain containing 4 |
| chr19_-_7040190 | 0.48 |
ENST00000381394.4
|
MBD3L4
|
methyl-CpG binding domain protein 3-like 4 |
| chr7_+_23637763 | 0.48 |
ENST00000410069.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr2_+_120770645 | 0.48 |
ENST00000443902.2
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr4_+_89299885 | 0.48 |
ENST00000380265.5
ENST00000273960.3 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr8_+_38585704 | 0.47 |
ENST00000519416.1
ENST00000520615.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr15_+_74165945 | 0.45 |
ENST00000535547.2
ENST00000300504.2 ENST00000562056.1 |
TBC1D21
|
TBC1 domain family, member 21 |
| chrX_+_7810303 | 0.45 |
ENST00000381059.3
ENST00000341408.4 |
VCX
|
variable charge, X-linked |
| chr2_+_101591314 | 0.44 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr12_-_53074182 | 0.44 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr4_+_89299994 | 0.43 |
ENST00000264346.7
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr1_+_43855560 | 0.43 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chrX_+_108779004 | 0.42 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr8_+_101170257 | 0.42 |
ENST00000251809.3
|
SPAG1
|
sperm associated antigen 1 |
| chr12_-_10251603 | 0.42 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr19_+_11658655 | 0.42 |
ENST00000588935.1
|
CNN1
|
calponin 1, basic, smooth muscle |
| chr1_+_160765884 | 0.41 |
ENST00000392203.4
|
LY9
|
lymphocyte antigen 9 |
| chr19_+_56989609 | 0.41 |
ENST00000601875.1
|
ZNF667-AS1
|
ZNF667 antisense RNA 1 (head to head) |
| chr1_+_160765860 | 0.40 |
ENST00000368037.5
|
LY9
|
lymphocyte antigen 9 |
| chr1_-_85100703 | 0.40 |
ENST00000370624.1
|
C1orf180
|
chromosome 1 open reading frame 180 |
| chr1_+_45212074 | 0.39 |
ENST00000372217.1
|
KIF2C
|
kinesin family member 2C |
| chr3_-_150690471 | 0.39 |
ENST00000468836.1
ENST00000328863.4 |
CLRN1
|
clarin 1 |
| chr4_+_68424434 | 0.39 |
ENST00000265404.2
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr12_-_10251576 | 0.39 |
ENST00000315330.4
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr15_+_66797455 | 0.39 |
ENST00000446801.2
|
ZWILCH
|
zwilch kinetochore protein |
| chr1_+_160765919 | 0.38 |
ENST00000341032.4
ENST00000368041.2 ENST00000368040.1 |
LY9
|
lymphocyte antigen 9 |
| chr9_-_130679257 | 0.37 |
ENST00000361444.3
ENST00000335791.5 ENST00000343609.2 |
ST6GALNAC4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr3_+_157154578 | 0.37 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr7_+_108210012 | 0.36 |
ENST00000249356.3
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr3_+_128444994 | 0.35 |
ENST00000482525.1
|
RAB7A
|
RAB7A, member RAS oncogene family |
| chr2_+_210517895 | 0.34 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr17_-_56494713 | 0.33 |
ENST00000407977.2
|
RNF43
|
ring finger protein 43 |
| chr14_+_88471468 | 0.33 |
ENST00000267549.3
|
GPR65
|
G protein-coupled receptor 65 |
| chr8_-_33424636 | 0.33 |
ENST00000256257.1
|
RNF122
|
ring finger protein 122 |
| chr9_-_99417562 | 0.33 |
ENST00000375234.3
ENST00000446045.1 |
AAED1
|
AhpC/TSA antioxidant enzyme domain containing 1 |
| chr8_+_30244580 | 0.32 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr11_+_6226782 | 0.32 |
ENST00000316375.2
|
C11orf42
|
chromosome 11 open reading frame 42 |
| chr17_-_56494882 | 0.32 |
ENST00000584437.1
|
RNF43
|
ring finger protein 43 |
| chr4_-_140544386 | 0.31 |
ENST00000561977.1
|
RP11-308D13.3
|
RP11-308D13.3 |
| chr6_+_31462658 | 0.31 |
ENST00000538442.1
|
MICB
|
MHC class I polypeptide-related sequence B |
| chr18_+_32455201 | 0.31 |
ENST00000590831.2
|
DTNA
|
dystrobrevin, alpha |
| chr1_-_43855479 | 0.31 |
ENST00000290663.6
ENST00000372457.4 |
MED8
|
mediator complex subunit 8 |
| chr16_-_30440530 | 0.31 |
ENST00000568434.1
|
DCTPP1
|
dCTP pyrophosphatase 1 |
| chr1_-_27952741 | 0.31 |
ENST00000399173.1
|
FGR
|
feline Gardner-Rasheed sarcoma viral oncogene homolog |
| chr17_-_27054952 | 0.31 |
ENST00000580518.1
|
TLCD1
|
TLC domain containing 1 |
| chr15_-_55881135 | 0.30 |
ENST00000302000.6
|
PYGO1
|
pygopus family PHD finger 1 |
| chrX_-_30877837 | 0.29 |
ENST00000378930.3
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr7_-_33140498 | 0.27 |
ENST00000448915.1
|
RP9
|
retinitis pigmentosa 9 (autosomal dominant) |
| chr12_-_92536433 | 0.27 |
ENST00000551563.2
ENST00000546975.1 ENST00000549802.1 |
C12orf79
|
chromosome 12 open reading frame 79 |
| chr12_+_108523133 | 0.27 |
ENST00000547525.1
|
WSCD2
|
WSC domain containing 2 |
| chr10_-_111713633 | 0.27 |
ENST00000538668.1
ENST00000369657.1 ENST00000369655.1 |
RP11-451M19.3
|
Uncharacterized protein |
| chr17_-_56494908 | 0.26 |
ENST00000577716.1
|
RNF43
|
ring finger protein 43 |
| chr3_-_57233966 | 0.26 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr4_-_68749745 | 0.26 |
ENST00000283916.6
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr2_-_85641162 | 0.26 |
ENST00000447219.2
ENST00000409670.1 ENST00000409724.1 |
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr17_+_66511540 | 0.25 |
ENST00000588188.2
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr7_+_99425633 | 0.25 |
ENST00000354829.2
ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chr4_-_68749699 | 0.24 |
ENST00000545541.1
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr9_+_5890802 | 0.24 |
ENST00000381477.3
ENST00000381476.1 ENST00000381471.1 |
MLANA
|
melan-A |
| chr9_+_108424738 | 0.24 |
ENST00000334077.3
|
TAL2
|
T-cell acute lymphocytic leukemia 2 |
| chr2_+_120770686 | 0.23 |
ENST00000331393.4
ENST00000443124.1 |
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr11_+_63137251 | 0.23 |
ENST00000310969.4
ENST00000279178.3 |
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9 |
| chr1_+_222988464 | 0.23 |
ENST00000420335.1
|
RP11-452F19.3
|
RP11-452F19.3 |
| chr6_-_26124138 | 0.23 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chrX_+_102024075 | 0.23 |
ENST00000431616.1
ENST00000440496.1 ENST00000420471.1 ENST00000435966.1 |
LINC00630
|
long intergenic non-protein coding RNA 630 |
| chr14_+_52456327 | 0.22 |
ENST00000556760.1
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr17_+_17685422 | 0.22 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr1_+_239882842 | 0.22 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor, muscarinic 3 |
| chr5_-_110062384 | 0.22 |
ENST00000429839.2
|
TMEM232
|
transmembrane protein 232 |
| chr3_+_128444965 | 0.22 |
ENST00000265062.3
|
RAB7A
|
RAB7A, member RAS oncogene family |
| chr10_+_105005644 | 0.22 |
ENST00000441178.2
|
RP11-332O19.5
|
ribulose-5-phosphate-3-epimerase-like 1 |
| chr3_+_63428752 | 0.22 |
ENST00000295894.5
|
SYNPR
|
synaptoporin |
| chr13_+_30510003 | 0.22 |
ENST00000400540.1
|
LINC00544
|
long intergenic non-protein coding RNA 544 |
| chr1_+_36335351 | 0.22 |
ENST00000373206.1
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr5_-_110062349 | 0.21 |
ENST00000511883.2
ENST00000455884.2 |
TMEM232
|
transmembrane protein 232 |
| chr18_+_44812072 | 0.21 |
ENST00000598649.1
ENST00000586905.2 |
CTD-2130O13.1
|
CTD-2130O13.1 |
| chr10_+_60936347 | 0.21 |
ENST00000373880.4
|
PHYHIPL
|
phytanoyl-CoA 2-hydroxylase interacting protein-like |
| chr6_-_130543958 | 0.20 |
ENST00000437477.2
ENST00000439090.2 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr4_-_123542224 | 0.19 |
ENST00000264497.3
|
IL21
|
interleukin 21 |
| chr12_+_123949053 | 0.19 |
ENST00000350887.5
|
SNRNP35
|
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr19_+_49866331 | 0.19 |
ENST00000597873.1
|
DKKL1
|
dickkopf-like 1 |
| chr11_-_5537920 | 0.19 |
ENST00000380184.1
|
UBQLNL
|
ubiquilin-like |
| chr2_+_132160448 | 0.19 |
ENST00000437751.1
|
AC073869.19
|
long intergenic non-protein coding RNA 1120 |
| chr19_+_54619125 | 0.18 |
ENST00000445811.1
ENST00000419967.1 ENST00000445124.1 ENST00000447810.1 |
PRPF31
|
pre-mRNA processing factor 31 |
| chr3_+_57875711 | 0.18 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chr1_+_117963209 | 0.17 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr1_-_27701307 | 0.17 |
ENST00000270879.4
ENST00000354982.2 |
FCN3
|
ficolin (collagen/fibrinogen domain containing) 3 |
| chr16_+_5121814 | 0.17 |
ENST00000262374.5
ENST00000586840.1 |
ALG1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr1_+_248402231 | 0.17 |
ENST00000306687.1
|
OR2M4
|
olfactory receptor, family 2, subfamily M, member 4 |
| chr6_-_10747802 | 0.17 |
ENST00000606522.1
ENST00000606652.1 |
RP11-421M1.8
|
RP11-421M1.8 |
| chr1_+_146373546 | 0.17 |
ENST00000446760.2
|
NBPF12
|
neuroblastoma breakpoint family, member 12 |
| chr12_-_102874378 | 0.17 |
ENST00000456098.1
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr19_+_56989485 | 0.16 |
ENST00000585445.1
ENST00000586091.1 ENST00000594783.1 ENST00000592146.1 ENST00000588158.1 ENST00000299997.4 ENST00000591797.1 |
ZNF667-AS1
|
ZNF667 antisense RNA 1 (head to head) |
| chr9_-_26892765 | 0.16 |
ENST00000520187.1
ENST00000333916.5 |
CAAP1
|
caspase activity and apoptosis inhibitor 1 |
| chr5_-_34919094 | 0.16 |
ENST00000341754.4
|
RAD1
|
RAD1 homolog (S. pombe) |
| chr3_+_132036207 | 0.15 |
ENST00000336375.5
ENST00000495911.1 |
ACPP
|
acid phosphatase, prostate |
| chr15_-_34628951 | 0.15 |
ENST00000397707.2
ENST00000560611.1 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr20_+_36373032 | 0.15 |
ENST00000373473.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr4_-_38806404 | 0.15 |
ENST00000308979.2
ENST00000505940.1 ENST00000515861.1 |
TLR1
|
toll-like receptor 1 |
| chr14_+_52456193 | 0.15 |
ENST00000261700.3
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr11_-_125550764 | 0.14 |
ENST00000527795.1
|
ACRV1
|
acrosomal vesicle protein 1 |
| chr13_-_38172863 | 0.14 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr2_-_206950781 | 0.14 |
ENST00000403263.1
|
INO80D
|
INO80 complex subunit D |
| chr12_-_7596735 | 0.14 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr11_-_57417405 | 0.14 |
ENST00000524669.1
ENST00000300022.3 |
YPEL4
|
yippee-like 4 (Drosophila) |
| chr7_+_132937820 | 0.14 |
ENST00000393161.2
ENST00000253861.4 |
EXOC4
|
exocyst complex component 4 |
| chr6_+_33422343 | 0.14 |
ENST00000395064.2
|
ZBTB9
|
zinc finger and BTB domain containing 9 |
| chr10_-_105992059 | 0.14 |
ENST00000369720.1
ENST00000369719.1 ENST00000357060.3 ENST00000428666.1 ENST00000278064.2 |
WDR96
|
WD repeat domain 96 |
| chr9_-_136006496 | 0.14 |
ENST00000372062.3
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr12_-_8815215 | 0.14 |
ENST00000544889.1
ENST00000543369.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr12_-_102874416 | 0.13 |
ENST00000392904.1
ENST00000337514.6 |
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr3_+_122103014 | 0.13 |
ENST00000232125.5
ENST00000477892.1 ENST00000469967.1 |
FAM162A
|
family with sequence similarity 162, member A |
| chr3_-_49131788 | 0.13 |
ENST00000395443.2
ENST00000411682.1 |
QRICH1
|
glutamine-rich 1 |
| chr2_+_90121477 | 0.13 |
ENST00000483379.1
|
IGKV1D-17
|
immunoglobulin kappa variable 1D-17 |
| chr6_-_52628271 | 0.13 |
ENST00000493422.1
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr11_+_118398178 | 0.13 |
ENST00000302783.4
ENST00000539546.1 |
TTC36
|
tetratricopeptide repeat domain 36 |
| chr15_+_64428529 | 0.12 |
ENST00000560861.1
|
SNX1
|
sorting nexin 1 |
| chr14_+_90863327 | 0.12 |
ENST00000356978.4
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr1_+_162039558 | 0.11 |
ENST00000530878.1
ENST00000361897.5 |
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein |
| chr13_+_43355732 | 0.11 |
ENST00000313851.1
|
FAM216B
|
family with sequence similarity 216, member B |
| chr6_+_31082603 | 0.11 |
ENST00000259881.9
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr1_-_19811132 | 0.11 |
ENST00000433834.1
|
CAPZB
|
capping protein (actin filament) muscle Z-line, beta |
| chr1_-_230513367 | 0.11 |
ENST00000321327.2
ENST00000525115.1 |
PGBD5
|
piggyBac transposable element derived 5 |
| chr12_+_21679220 | 0.10 |
ENST00000256969.2
|
C12orf39
|
chromosome 12 open reading frame 39 |
| chr16_+_72459838 | 0.10 |
ENST00000564508.1
|
AC004158.3
|
AC004158.3 |
| chr3_+_57875738 | 0.10 |
ENST00000417128.1
ENST00000438794.1 |
SLMAP
|
sarcolemma associated protein |
| chr9_+_133569108 | 0.10 |
ENST00000372358.5
ENST00000546165.1 ENST00000372352.3 ENST00000372351.3 ENST00000372350.3 ENST00000495699.2 |
EXOSC2
|
exosome component 2 |
| chr3_+_52017454 | 0.10 |
ENST00000476854.1
ENST00000476351.1 ENST00000494103.1 ENST00000404366.2 ENST00000469863.1 |
ACY1
|
aminoacylase 1 |
| chr17_-_79838860 | 0.09 |
ENST00000582866.1
|
RP11-498C9.15
|
RP11-498C9.15 |
| chr12_-_8815299 | 0.09 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr14_+_37126765 | 0.09 |
ENST00000402703.2
|
PAX9
|
paired box 9 |
| chr12_+_27863706 | 0.09 |
ENST00000081029.3
ENST00000538315.1 ENST00000542791.1 |
MRPS35
|
mitochondrial ribosomal protein S35 |
| chr7_+_72742178 | 0.09 |
ENST00000442793.1
ENST00000413573.2 ENST00000252037.4 |
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr16_+_3068393 | 0.09 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chrX_+_22056165 | 0.08 |
ENST00000535894.1
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr22_-_39268308 | 0.08 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr3_+_178866199 | 0.08 |
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr6_-_167571817 | 0.08 |
ENST00000366834.1
|
GPR31
|
G protein-coupled receptor 31 |
| chr20_+_55926583 | 0.08 |
ENST00000395840.2
|
RAE1
|
ribonucleic acid export 1 |
| chr8_+_85095769 | 0.07 |
ENST00000518566.1
|
RALYL
|
RALY RNA binding protein-like |
| chr10_+_50887683 | 0.07 |
ENST00000374113.3
ENST00000374111.3 ENST00000374112.3 ENST00000535836.1 |
C10orf53
|
chromosome 10 open reading frame 53 |
| chr12_-_57941004 | 0.07 |
ENST00000550750.1
ENST00000548249.1 |
DCTN2
|
dynactin 2 (p50) |
| chr3_-_109056419 | 0.07 |
ENST00000335658.6
|
DPPA4
|
developmental pluripotency associated 4 |
| chr12_+_6833237 | 0.07 |
ENST00000229251.3
ENST00000539735.1 ENST00000538410.1 |
COPS7A
|
COP9 signalosome subunit 7A |
| chr1_+_32739733 | 0.07 |
ENST00000333070.4
|
LCK
|
lymphocyte-specific protein tyrosine kinase |
| chr14_+_38264418 | 0.06 |
ENST00000267368.7
ENST00000382320.3 |
TTC6
|
tetratricopeptide repeat domain 6 |
| chr17_-_64225508 | 0.06 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr3_-_64009658 | 0.06 |
ENST00000394431.2
|
PSMD6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr1_-_38019878 | 0.06 |
ENST00000296215.6
|
SNIP1
|
Smad nuclear interacting protein 1 |
| chr4_-_159956333 | 0.06 |
ENST00000434826.2
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr1_+_73771844 | 0.06 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
| chr6_+_10747986 | 0.06 |
ENST00000379542.5
|
TMEM14B
|
transmembrane protein 14B |
| chr18_+_3447572 | 0.05 |
ENST00000548489.2
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr2_-_74619152 | 0.05 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chr10_+_28822236 | 0.05 |
ENST00000347934.4
ENST00000354911.4 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr4_+_57845024 | 0.05 |
ENST00000431623.2
ENST00000441246.2 |
POLR2B
|
polymerase (RNA) II (DNA directed) polypeptide B, 140kDa |
| chr17_+_73629500 | 0.05 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr7_-_144435985 | 0.05 |
ENST00000549981.1
|
TPK1
|
thiamin pyrophosphokinase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXJ2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.8 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.8 | 4.5 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.4 | 1.7 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.4 | 1.6 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.4 | 1.1 | GO:2001190 | natural killer cell tolerance induction(GO:0002519) positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.3 | 2.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.2 | 1.7 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.2 | 1.0 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.2 | 3.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.2 | 0.6 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.2 | 0.9 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 0.8 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 0.6 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.4 | GO:1903973 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.1 | 1.1 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 0.3 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 1.7 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.1 | 1.4 | GO:0032740 | positive regulation of interleukin-17 production(GO:0032740) |
| 0.1 | 0.5 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.3 | GO:0009149 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.1 | 0.4 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.1 | 0.2 | GO:0051598 | meiotic recombination checkpoint(GO:0051598) |
| 0.0 | 0.1 | GO:0070340 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.7 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.3 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:1903947 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.0 | 0.7 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.2 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.2 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 1.2 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 1.0 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.2 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) bone regeneration(GO:1990523) |
| 0.0 | 0.1 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.4 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.4 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.7 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.3 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.3 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.5 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.3 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.0 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.4 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.3 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 1.7 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.2 | 1.0 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.2 | 0.7 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.8 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.1 | 0.5 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.6 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 0.3 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.3 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.4 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 1.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.8 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.8 | 4.5 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 1.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.2 | 1.7 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 1.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.8 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.5 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.7 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.3 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 3.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 2.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.4 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.3 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 1.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.4 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.5 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.3 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 2.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 3.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 4.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 3.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 1.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.1 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 1.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |