Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for FOXN1
Z-value: 0.66
Transcription factors associated with FOXN1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXN1
|
ENSG00000109101.3 | forkhead box N1 |
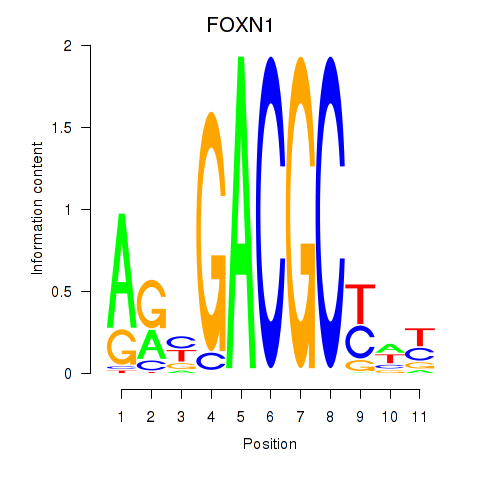
Activity profile of FOXN1 motif
Sorted Z-values of FOXN1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_82266053 | 1.36 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr20_+_30193083 | 1.32 |
ENST00000376112.3
ENST00000376105.3 |
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein |
| chr1_+_221054584 | 1.31 |
ENST00000549319.1
|
HLX
|
H2.0-like homeobox |
| chr7_-_120498357 | 1.23 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr17_+_40913264 | 1.12 |
ENST00000587142.1
ENST00000588576.1 |
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr10_-_125851961 | 1.11 |
ENST00000346248.5
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
| chr7_+_89841000 | 1.05 |
ENST00000287908.3
|
STEAP2
|
STEAP family member 2, metalloreductase |
| chr17_+_40912764 | 1.04 |
ENST00000589683.1
ENST00000588928.1 |
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr7_+_89841024 | 1.02 |
ENST00000394626.1
|
STEAP2
|
STEAP family member 2, metalloreductase |
| chr17_-_58469591 | 1.01 |
ENST00000589335.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr17_+_40913210 | 0.93 |
ENST00000253796.5
|
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr4_+_42399856 | 0.90 |
ENST00000319234.4
|
SHISA3
|
shisa family member 3 |
| chr5_-_100238956 | 0.80 |
ENST00000231461.5
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr20_+_58515417 | 0.76 |
ENST00000360816.3
|
FAM217B
|
family with sequence similarity 217, member B |
| chrX_+_153237740 | 0.75 |
ENST00000369982.4
|
TMEM187
|
transmembrane protein 187 |
| chr14_+_61788429 | 0.73 |
ENST00000332981.5
|
PRKCH
|
protein kinase C, eta |
| chrX_-_117107680 | 0.72 |
ENST00000447671.2
ENST00000262820.3 |
KLHL13
|
kelch-like family member 13 |
| chr20_-_36156125 | 0.69 |
ENST00000397135.1
ENST00000397137.1 |
BLCAP
|
bladder cancer associated protein |
| chr17_-_64187973 | 0.67 |
ENST00000583358.1
ENST00000392769.2 |
CEP112
|
centrosomal protein 112kDa |
| chrX_-_117107542 | 0.63 |
ENST00000371878.1
|
KLHL13
|
kelch-like family member 13 |
| chr14_-_77495007 | 0.60 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr3_+_52280220 | 0.59 |
ENST00000409502.3
ENST00000323588.4 |
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr12_+_12938541 | 0.59 |
ENST00000356591.4
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr2_-_152684977 | 0.58 |
ENST00000428992.2
ENST00000295087.8 |
ARL5A
|
ADP-ribosylation factor-like 5A |
| chr8_-_11058847 | 0.57 |
ENST00000297303.4
ENST00000416569.2 |
XKR6
|
XK, Kell blood group complex subunit-related family, member 6 |
| chr13_-_110438914 | 0.57 |
ENST00000375856.3
|
IRS2
|
insulin receptor substrate 2 |
| chr2_+_73612858 | 0.54 |
ENST00000409009.1
ENST00000264448.6 ENST00000377715.1 |
ALMS1
|
Alstrom syndrome 1 |
| chr3_+_52280173 | 0.54 |
ENST00000296487.4
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr12_+_51632666 | 0.53 |
ENST00000604900.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr2_+_113403434 | 0.53 |
ENST00000272542.3
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1 |
| chr11_-_82782861 | 0.50 |
ENST00000524635.1
ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30
|
RAB30, member RAS oncogene family |
| chr15_+_92937058 | 0.48 |
ENST00000268164.3
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr12_+_32260085 | 0.48 |
ENST00000548411.1
ENST00000281474.5 ENST00000551086.1 |
BICD1
|
bicaudal D homolog 1 (Drosophila) |
| chr15_+_41221536 | 0.47 |
ENST00000249749.5
|
DLL4
|
delta-like 4 (Drosophila) |
| chr17_+_44928946 | 0.46 |
ENST00000290015.2
ENST00000393461.2 |
WNT9B
|
wingless-type MMTV integration site family, member 9B |
| chr10_-_52383644 | 0.46 |
ENST00000361781.2
|
SGMS1
|
sphingomyelin synthase 1 |
| chr19_-_14640005 | 0.44 |
ENST00000596853.1
ENST00000596075.1 ENST00000595992.1 ENST00000396969.4 ENST00000601533.1 ENST00000598692.1 |
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr5_-_131892501 | 0.43 |
ENST00000450655.1
|
IL5
|
interleukin 5 (colony-stimulating factor, eosinophil) |
| chrX_-_106959631 | 0.43 |
ENST00000486554.1
ENST00000372390.4 |
TSC22D3
|
TSC22 domain family, member 3 |
| chrX_+_51928002 | 0.42 |
ENST00000375626.3
|
MAGED4
|
melanoma antigen family D, 4 |
| chr13_+_27131798 | 0.41 |
ENST00000361042.4
|
WASF3
|
WAS protein family, member 3 |
| chr20_-_36156293 | 0.41 |
ENST00000373537.2
ENST00000414542.2 |
BLCAP
|
bladder cancer associated protein |
| chr4_-_170533723 | 0.40 |
ENST00000510533.1
ENST00000439128.2 ENST00000511633.1 ENST00000512193.1 ENST00000507142.1 |
NEK1
|
NIMA-related kinase 1 |
| chr5_+_172261228 | 0.39 |
ENST00000393784.3
|
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr17_-_64188177 | 0.39 |
ENST00000535342.2
|
CEP112
|
centrosomal protein 112kDa |
| chr7_-_132261253 | 0.39 |
ENST00000321063.4
|
PLXNA4
|
plexin A4 |
| chr17_-_58469687 | 0.39 |
ENST00000590133.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr13_+_27131887 | 0.38 |
ENST00000335327.5
|
WASF3
|
WAS protein family, member 3 |
| chr15_-_55700216 | 0.38 |
ENST00000569205.1
|
CCPG1
|
cell cycle progression 1 |
| chr2_-_68694390 | 0.38 |
ENST00000377957.3
|
FBXO48
|
F-box protein 48 |
| chr7_-_132262060 | 0.38 |
ENST00000359827.3
|
PLXNA4
|
plexin A4 |
| chr19_-_37663332 | 0.37 |
ENST00000392157.2
|
ZNF585A
|
zinc finger protein 585A |
| chr15_+_83776137 | 0.37 |
ENST00000322019.9
|
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr1_-_59012365 | 0.36 |
ENST00000456980.1
ENST00000482274.2 ENST00000453710.1 ENST00000419242.1 ENST00000358603.2 ENST00000371226.3 ENST00000426139.1 |
OMA1
|
OMA1 zinc metallopeptidase |
| chr15_-_55700457 | 0.35 |
ENST00000442196.3
ENST00000563171.1 ENST00000425574.3 |
CCPG1
|
cell cycle progression 1 |
| chr6_+_7590413 | 0.35 |
ENST00000342415.5
|
SNRNP48
|
small nuclear ribonucleoprotein 48kDa (U11/U12) |
| chr19_-_37663572 | 0.35 |
ENST00000588354.1
ENST00000292841.5 ENST00000355533.2 ENST00000356958.4 |
ZNF585A
|
zinc finger protein 585A |
| chr5_+_99871004 | 0.34 |
ENST00000312637.4
|
FAM174A
|
family with sequence similarity 174, member A |
| chr15_+_55700741 | 0.34 |
ENST00000569691.1
|
C15orf65
|
chromosome 15 open reading frame 65 |
| chr15_+_83776324 | 0.33 |
ENST00000379390.6
ENST00000379386.4 ENST00000565774.1 ENST00000565982.1 |
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr2_-_28113217 | 0.33 |
ENST00000444339.2
|
RBKS
|
ribokinase |
| chr2_+_54198210 | 0.32 |
ENST00000607452.1
ENST00000422521.2 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr5_-_93447333 | 0.32 |
ENST00000395965.3
ENST00000505869.1 ENST00000509163.1 |
FAM172A
|
family with sequence similarity 172, member A |
| chr15_-_55700522 | 0.32 |
ENST00000564092.1
ENST00000310958.6 |
CCPG1
|
cell cycle progression 1 |
| chr19_-_47164386 | 0.31 |
ENST00000391916.2
ENST00000410105.2 |
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr6_-_119670919 | 0.31 |
ENST00000368468.3
|
MAN1A1
|
mannosidase, alpha, class 1A, member 1 |
| chr3_-_133380731 | 0.31 |
ENST00000260810.5
|
TOPBP1
|
topoisomerase (DNA) II binding protein 1 |
| chr2_-_128785688 | 0.31 |
ENST00000259234.6
|
SAP130
|
Sin3A-associated protein, 130kDa |
| chr2_-_38830030 | 0.30 |
ENST00000410076.1
|
HNRNPLL
|
heterogeneous nuclear ribonucleoprotein L-like |
| chr1_+_74663896 | 0.29 |
ENST00000370898.3
ENST00000467578.2 ENST00000370894.5 ENST00000482102.2 ENST00000609362.1 ENST00000534056.1 ENST00000557284.2 ENST00000370899.3 ENST00000370895.1 ENST00000534632.1 ENST00000370893.1 ENST00000370891.2 |
FPGT
FPGT-TNNI3K
TNNI3K
|
fucose-1-phosphate guanylyltransferase FPGT-TNNI3K readthrough TNNI3 interacting kinase |
| chr7_+_138916231 | 0.29 |
ENST00000473989.3
ENST00000288561.8 |
UBN2
|
ubinuclein 2 |
| chr1_-_114301755 | 0.28 |
ENST00000393357.2
ENST00000369596.2 ENST00000446739.1 |
PHTF1
|
putative homeodomain transcription factor 1 |
| chr17_-_76921459 | 0.28 |
ENST00000262768.7
|
TIMP2
|
TIMP metallopeptidase inhibitor 2 |
| chr1_-_114301960 | 0.28 |
ENST00000369598.1
ENST00000369600.1 |
PHTF1
|
putative homeodomain transcription factor 1 |
| chr5_+_122847908 | 0.28 |
ENST00000511130.2
ENST00000512718.3 |
CSNK1G3
|
casein kinase 1, gamma 3 |
| chr3_+_49977490 | 0.28 |
ENST00000539992.1
|
RBM6
|
RNA binding motif protein 6 |
| chr4_+_148538517 | 0.28 |
ENST00000296582.3
ENST00000508208.1 |
TMEM184C
|
transmembrane protein 184C |
| chr7_+_36192855 | 0.27 |
ENST00000534978.1
|
EEPD1
|
endonuclease/exonuclease/phosphatase family domain containing 1 |
| chr17_-_7387524 | 0.27 |
ENST00000311403.4
|
ZBTB4
|
zinc finger and BTB domain containing 4 |
| chr11_-_65325430 | 0.27 |
ENST00000322147.4
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr1_-_78149041 | 0.27 |
ENST00000414381.1
ENST00000370798.1 |
ZZZ3
|
zinc finger, ZZ-type containing 3 |
| chr12_+_57943781 | 0.27 |
ENST00000455537.2
ENST00000286452.5 |
KIF5A
|
kinesin family member 5A |
| chr1_+_245133656 | 0.27 |
ENST00000366521.3
|
EFCAB2
|
EF-hand calcium binding domain 2 |
| chr4_-_54457783 | 0.26 |
ENST00000263925.7
ENST00000512247.1 |
LNX1
|
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
| chr3_+_49977523 | 0.26 |
ENST00000422955.1
|
RBM6
|
RNA binding motif protein 6 |
| chr20_-_58515344 | 0.26 |
ENST00000370996.3
|
PPP1R3D
|
protein phosphatase 1, regulatory subunit 3D |
| chr5_+_61602055 | 0.26 |
ENST00000381103.2
|
KIF2A
|
kinesin heavy chain member 2A |
| chr17_+_7387677 | 0.25 |
ENST00000322644.6
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr12_+_56473910 | 0.25 |
ENST00000411731.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr10_-_104179682 | 0.25 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr1_-_114302086 | 0.25 |
ENST00000369604.1
ENST00000357783.2 |
PHTF1
|
putative homeodomain transcription factor 1 |
| chr19_-_46389359 | 0.25 |
ENST00000302165.3
|
IRF2BP1
|
interferon regulatory factor 2 binding protein 1 |
| chr5_+_154237778 | 0.25 |
ENST00000523698.1
ENST00000517876.1 ENST00000520472.1 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr5_+_93954039 | 0.25 |
ENST00000265140.5
|
ANKRD32
|
ankyrin repeat domain 32 |
| chr20_-_3154162 | 0.25 |
ENST00000360342.3
|
LZTS3
|
Homo sapiens leucine zipper, putative tumor suppressor family member 3 (LZTS3), mRNA. |
| chr1_+_206138457 | 0.25 |
ENST00000367128.3
ENST00000431655.2 |
FAM72A
|
family with sequence similarity 72, member A |
| chr17_-_33905521 | 0.24 |
ENST00000225873.4
|
PEX12
|
peroxisomal biogenesis factor 12 |
| chr2_+_25264933 | 0.24 |
ENST00000401432.3
ENST00000403714.3 |
EFR3B
|
EFR3 homolog B (S. cerevisiae) |
| chr3_+_137906109 | 0.24 |
ENST00000481646.1
ENST00000469044.1 ENST00000491704.1 ENST00000461600.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr8_-_38126635 | 0.24 |
ENST00000529359.1
|
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr4_+_128651530 | 0.24 |
ENST00000281154.4
|
SLC25A31
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31 |
| chr12_-_88535842 | 0.24 |
ENST00000550962.1
ENST00000552810.1 |
CEP290
|
centrosomal protein 290kDa |
| chr16_-_30773372 | 0.23 |
ENST00000545825.1
ENST00000541260.1 |
C16orf93
|
chromosome 16 open reading frame 93 |
| chr17_+_62075703 | 0.23 |
ENST00000577953.1
ENST00000582540.1 ENST00000579184.1 ENST00000425164.3 ENST00000412177.1 ENST00000539996.1 ENST00000583891.1 ENST00000580752.1 |
C17orf72
|
chromosome 17 open reading frame 72 |
| chr7_+_131012605 | 0.23 |
ENST00000446815.1
ENST00000352689.6 |
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr18_-_72921303 | 0.23 |
ENST00000322342.3
|
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr5_+_154238042 | 0.22 |
ENST00000519211.1
ENST00000522458.1 ENST00000519903.1 ENST00000521450.1 ENST00000403027.2 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr11_-_66360548 | 0.22 |
ENST00000333861.3
|
CCDC87
|
coiled-coil domain containing 87 |
| chr14_+_96829886 | 0.22 |
ENST00000556095.1
|
GSKIP
|
GSK3B interacting protein |
| chr7_+_96747030 | 0.22 |
ENST00000360382.4
|
ACN9
|
ACN9 homolog (S. cerevisiae) |
| chr19_-_48867291 | 0.22 |
ENST00000435956.3
|
TMEM143
|
transmembrane protein 143 |
| chr2_-_38830090 | 0.22 |
ENST00000449105.3
|
HNRNPLL
|
heterogeneous nuclear ribonucleoprotein L-like |
| chr7_-_92157760 | 0.21 |
ENST00000248633.4
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr2_-_73520667 | 0.21 |
ENST00000545030.1
ENST00000436467.2 |
EGR4
|
early growth response 4 |
| chr6_+_52930237 | 0.21 |
ENST00000323557.7
|
FBXO9
|
F-box protein 9 |
| chr19_-_19144243 | 0.21 |
ENST00000594445.1
ENST00000452918.2 ENST00000600377.1 ENST00000337018.6 |
SUGP2
|
SURP and G patch domain containing 2 |
| chr20_+_62669472 | 0.21 |
ENST00000463337.1
ENST00000358393.1 |
LINC00176
|
long intergenic non-protein coding RNA 176 |
| chr1_-_143913143 | 0.21 |
ENST00000400889.1
|
FAM72D
|
family with sequence similarity 72, member D |
| chr3_+_47021168 | 0.21 |
ENST00000450053.3
ENST00000292309.5 ENST00000383740.2 |
NBEAL2
|
neurobeachin-like 2 |
| chrX_-_99891796 | 0.21 |
ENST00000373020.4
|
TSPAN6
|
tetraspanin 6 |
| chrX_+_130192216 | 0.20 |
ENST00000276211.5
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr7_-_132261223 | 0.20 |
ENST00000423507.2
|
PLXNA4
|
plexin A4 |
| chr5_+_110559784 | 0.20 |
ENST00000282356.4
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr19_-_48867171 | 0.20 |
ENST00000377431.2
ENST00000436660.2 ENST00000541566.1 |
TMEM143
|
transmembrane protein 143 |
| chr4_-_155412868 | 0.20 |
ENST00000339452.1
ENST00000443500.1 |
DCHS2
|
dachsous cadherin-related 2 |
| chr5_+_131993856 | 0.19 |
ENST00000304506.3
|
IL13
|
interleukin 13 |
| chr11_+_64059464 | 0.19 |
ENST00000394525.2
|
KCNK4
|
potassium channel, subfamily K, member 4 |
| chr1_+_11994715 | 0.19 |
ENST00000449038.1
ENST00000376369.3 ENST00000429000.2 ENST00000196061.4 |
PLOD1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr1_-_245134273 | 0.19 |
ENST00000607453.1
|
RP11-156E8.1
|
Uncharacterized protein |
| chr19_+_41860047 | 0.19 |
ENST00000604123.1
|
TMEM91
|
transmembrane protein 91 |
| chr18_+_3448455 | 0.19 |
ENST00000549780.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr9_+_131799213 | 0.19 |
ENST00000358369.4
ENST00000406926.2 ENST00000277475.5 ENST00000450073.1 |
FAM73B
|
family with sequence similarity 73, member B |
| chr19_-_19626838 | 0.19 |
ENST00000360913.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr9_+_37667978 | 0.19 |
ENST00000539465.1
|
FRMPD1
|
FERM and PDZ domain containing 1 |
| chr21_+_17102311 | 0.19 |
ENST00000285679.6
ENST00000351097.5 ENST00000285681.2 ENST00000400183.2 |
USP25
|
ubiquitin specific peptidase 25 |
| chr7_-_92157747 | 0.18 |
ENST00000428214.1
ENST00000438045.1 |
PEX1
|
peroxisomal biogenesis factor 1 |
| chr5_+_65440032 | 0.18 |
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr20_-_31172598 | 0.18 |
ENST00000201961.2
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr5_+_131892603 | 0.18 |
ENST00000378823.3
ENST00000265335.6 |
RAD50
|
RAD50 homolog (S. cerevisiae) |
| chr5_+_154238096 | 0.18 |
ENST00000517568.1
ENST00000524105.1 ENST00000285896.6 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr17_-_47308128 | 0.18 |
ENST00000413580.1
ENST00000511066.1 |
PHOSPHO1
|
phosphatase, orphan 1 |
| chr7_+_76751926 | 0.18 |
ENST00000285871.4
ENST00000431197.1 |
CCDC146
|
coiled-coil domain containing 146 |
| chr8_+_38243821 | 0.18 |
ENST00000519476.2
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr17_-_45266542 | 0.17 |
ENST00000531206.1
ENST00000527547.1 ENST00000446365.2 ENST00000575483.1 ENST00000066544.3 |
CDC27
|
cell division cycle 27 |
| chr14_+_96829814 | 0.17 |
ENST00000555181.1
ENST00000553699.1 ENST00000554182.1 |
GSKIP
|
GSK3B interacting protein |
| chr6_-_150039249 | 0.17 |
ENST00000543571.1
|
LATS1
|
large tumor suppressor kinase 1 |
| chr19_-_38146289 | 0.17 |
ENST00000392144.1
ENST00000591444.1 ENST00000351218.2 ENST00000587809.1 |
ZFP30
|
ZFP30 zinc finger protein |
| chr11_+_125774362 | 0.17 |
ENST00000530414.1
ENST00000530129.2 |
DDX25
|
DEAD (Asp-Glu-Ala-Asp) box helicase 25 |
| chr13_-_46626847 | 0.17 |
ENST00000242848.4
ENST00000282007.3 |
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chrX_+_101380642 | 0.17 |
ENST00000372780.1
ENST00000329035.2 |
TCEAL2
|
transcription elongation factor A (SII)-like 2 |
| chr1_-_114301503 | 0.17 |
ENST00000447664.2
|
PHTF1
|
putative homeodomain transcription factor 1 |
| chr7_-_148725733 | 0.17 |
ENST00000286091.4
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr14_-_92588013 | 0.17 |
ENST00000553514.1
ENST00000605997.1 |
NDUFB1
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa |
| chr5_-_1524015 | 0.17 |
ENST00000283415.3
|
LPCAT1
|
lysophosphatidylcholine acyltransferase 1 |
| chr10_+_93558069 | 0.16 |
ENST00000371627.4
|
TNKS2
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 |
| chr12_+_120933904 | 0.16 |
ENST00000550178.1
ENST00000550845.1 ENST00000549989.1 ENST00000552870.1 |
DYNLL1
|
dynein, light chain, LC8-type 1 |
| chr17_-_47308100 | 0.16 |
ENST00000503902.1
ENST00000512250.1 |
PHOSPHO1
|
phosphatase, orphan 1 |
| chr17_-_16395455 | 0.16 |
ENST00000409083.3
|
FAM211A
|
family with sequence similarity 211, member A |
| chr15_+_76135622 | 0.16 |
ENST00000338677.4
ENST00000267938.4 ENST00000569423.1 |
UBE2Q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr10_+_103113802 | 0.15 |
ENST00000370187.3
|
BTRC
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr12_+_49740700 | 0.15 |
ENST00000549441.2
ENST00000395069.3 |
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr2_-_20251744 | 0.15 |
ENST00000175091.4
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha |
| chr1_+_161195781 | 0.15 |
ENST00000367988.3
|
TOMM40L
|
translocase of outer mitochondrial membrane 40 homolog (yeast)-like |
| chr2_+_130939235 | 0.15 |
ENST00000425361.1
ENST00000457492.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr14_-_20929624 | 0.15 |
ENST00000398020.4
ENST00000250489.4 |
TMEM55B
|
transmembrane protein 55B |
| chr6_+_166945369 | 0.15 |
ENST00000598601.1
|
Z98049.1
|
CDNA FLJ25492 fis, clone CBR01389; Uncharacterized protein |
| chr10_+_18629628 | 0.15 |
ENST00000377329.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr14_+_93260569 | 0.15 |
ENST00000163416.2
|
GOLGA5
|
golgin A5 |
| chr19_-_9929708 | 0.15 |
ENST00000247977.4
ENST00000590277.1 ENST00000588922.1 ENST00000589626.1 ENST00000592067.1 ENST00000586469.1 |
FBXL12
|
F-box and leucine-rich repeat protein 12 |
| chr10_-_27529716 | 0.15 |
ENST00000375897.3
ENST00000396271.3 |
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr14_+_93260642 | 0.15 |
ENST00000355976.2
|
GOLGA5
|
golgin A5 |
| chr19_-_42806723 | 0.15 |
ENST00000262890.3
|
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr5_+_32710736 | 0.15 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr10_-_127408011 | 0.15 |
ENST00000531977.1
ENST00000527483.1 ENST00000525909.1 ENST00000528844.1 ENST00000423178.2 |
RP11-383C5.4
|
RP11-383C5.4 |
| chr4_+_110354928 | 0.14 |
ENST00000504968.2
ENST00000399100.2 ENST00000265175.5 |
SEC24B
|
SEC24 family member B |
| chr12_+_50451331 | 0.14 |
ENST00000228468.4
|
ASIC1
|
acid-sensing (proton-gated) ion channel 1 |
| chr5_+_154238149 | 0.14 |
ENST00000519430.1
ENST00000520671.1 ENST00000521583.1 ENST00000518028.1 ENST00000519404.1 ENST00000519394.1 ENST00000518775.1 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr12_+_120933859 | 0.14 |
ENST00000242577.6
ENST00000548214.1 ENST00000392508.2 |
DYNLL1
|
dynein, light chain, LC8-type 1 |
| chr11_+_125774258 | 0.14 |
ENST00000263576.6
|
DDX25
|
DEAD (Asp-Glu-Ala-Asp) box helicase 25 |
| chr17_-_40913275 | 0.14 |
ENST00000589716.1
ENST00000360166.3 |
RAMP2-AS1
|
RAMP2 antisense RNA 1 |
| chr14_+_54863739 | 0.14 |
ENST00000541304.1
|
CDKN3
|
cyclin-dependent kinase inhibitor 3 |
| chr2_+_46769798 | 0.14 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr10_-_119134918 | 0.14 |
ENST00000334464.5
|
PDZD8
|
PDZ domain containing 8 |
| chr15_-_45422056 | 0.14 |
ENST00000267803.4
ENST00000559014.1 ENST00000558851.1 ENST00000559988.1 ENST00000558996.1 ENST00000558422.1 ENST00000559226.1 ENST00000558326.1 ENST00000558377.1 ENST00000559644.1 |
DUOXA1
|
dual oxidase maturation factor 1 |
| chr10_+_14880157 | 0.13 |
ENST00000378372.3
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr15_+_66161792 | 0.13 |
ENST00000564910.1
ENST00000261890.2 |
RAB11A
|
RAB11A, member RAS oncogene family |
| chr9_+_91933407 | 0.13 |
ENST00000375807.3
ENST00000339901.4 |
SECISBP2
|
SECIS binding protein 2 |
| chr2_+_30670077 | 0.13 |
ENST00000466477.1
ENST00000465200.1 ENST00000379509.3 ENST00000319406.4 ENST00000488144.1 ENST00000465538.1 ENST00000309052.4 ENST00000359433.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr19_-_16770915 | 0.13 |
ENST00000593459.1
ENST00000358726.6 ENST00000597711.1 ENST00000487416.2 |
CTC-429P9.4
SMIM7
|
Small integral membrane protein 7; Uncharacterized protein small integral membrane protein 7 |
| chr3_-_195163803 | 0.13 |
ENST00000326793.6
|
ACAP2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr14_-_92588246 | 0.13 |
ENST00000329559.3
|
NDUFB1
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa |
| chr15_-_52472078 | 0.13 |
ENST00000396335.4
ENST00000560116.1 ENST00000358784.7 |
GNB5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr3_+_180319918 | 0.13 |
ENST00000296015.4
ENST00000491380.1 ENST00000412756.2 ENST00000382584.4 |
TTC14
|
tetratricopeptide repeat domain 14 |
| chr20_-_31124186 | 0.13 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr14_+_54863682 | 0.12 |
ENST00000543789.2
ENST00000442975.2 ENST00000458126.2 ENST00000556102.2 |
CDKN3
|
cyclin-dependent kinase inhibitor 3 |
| chr12_-_52967600 | 0.12 |
ENST00000549343.1
ENST00000305620.2 |
KRT74
|
keratin 74 |
| chr8_+_38243967 | 0.12 |
ENST00000524874.1
ENST00000379957.4 ENST00000523983.2 |
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr6_+_27215494 | 0.12 |
ENST00000230582.3
|
PRSS16
|
protease, serine, 16 (thymus) |
| chr15_+_66161802 | 0.12 |
ENST00000566233.1
ENST00000565075.1 ENST00000435304.2 |
RAB11A
|
RAB11A, member RAS oncogene family |
| chr17_-_40913029 | 0.12 |
ENST00000592195.1
ENST00000592670.1 ENST00000587694.1 ENST00000591082.1 |
RAMP2-AS1
|
RAMP2 antisense RNA 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXN1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.3 | 3.1 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.3 | 1.0 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.3 | 2.1 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.2 | 1.0 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 1.3 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.2 | 0.5 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.2 | 0.5 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.4 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.4 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.1 | 0.5 | GO:0072554 | blood vessel lumenization(GO:0072554) |
| 0.1 | 0.6 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 | 0.3 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.1 | 0.2 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.1 | 0.4 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.1 | 1.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.7 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 | 0.3 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.2 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.1 | 0.2 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.4 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.1 | 0.6 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 0.4 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 0.3 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.1 | 0.2 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 0.3 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.2 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.4 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.5 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.2 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.0 | 0.1 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.0 | 1.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.2 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 1.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.3 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 1.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.3 | GO:0048793 | pronephros development(GO:0048793) |
| 0.0 | 1.1 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0035625 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 0.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.8 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.1 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.3 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0098704 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.2 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 0.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 | 0.8 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.3 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.1 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.1 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.2 | 0.5 | GO:0039714 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.1 | 0.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 1.0 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.4 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 1.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 2.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.3 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 1.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.1 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.4 | 1.1 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.3 | 2.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.7 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.4 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 0.5 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 1.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 1.0 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.5 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.4 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 0.5 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.2 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.1 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 0.2 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.3 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 1.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.2 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.2 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 1.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.2 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 1.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.2 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.0 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.6 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.9 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 3.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.7 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |