Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
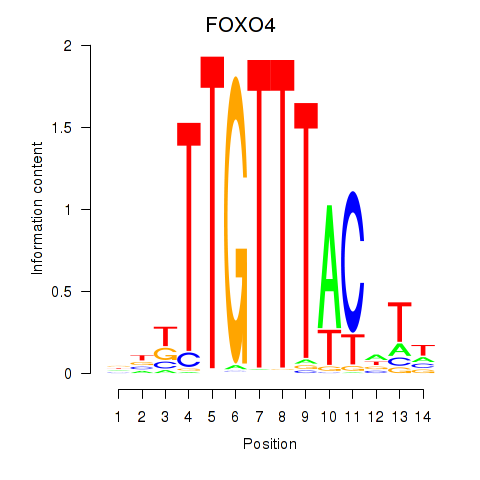
Results for FOXO4
Z-value: 0.90
Transcription factors associated with FOXO4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXO4
|
ENSG00000184481.12 | forkhead box O4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXO4 | hg19_v2_chrX_+_70316005_70316047 | 0.51 | 9.6e-03 | Click! |
Activity profile of FOXO4 motif
Sorted Z-values of FOXO4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_186696425 | 5.80 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_-_112127981 | 3.60 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr4_-_186697044 | 3.47 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr9_+_27109392 | 3.30 |
ENST00000406359.4
|
TEK
|
TEK tyrosine kinase, endothelial |
| chr2_+_12857015 | 3.00 |
ENST00000155926.4
|
TRIB2
|
tribbles pseudokinase 2 |
| chr2_+_12857043 | 3.00 |
ENST00000381465.2
|
TRIB2
|
tribbles pseudokinase 2 |
| chr4_-_70626314 | 2.88 |
ENST00000510821.1
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr13_+_32838801 | 2.87 |
ENST00000542859.1
|
FRY
|
furry homolog (Drosophila) |
| chr4_-_159080806 | 2.72 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr3_-_18466026 | 2.61 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr18_+_72922710 | 2.23 |
ENST00000322038.5
|
TSHZ1
|
teashirt zinc finger homeobox 1 |
| chr18_+_77160282 | 2.01 |
ENST00000318065.5
ENST00000545796.1 ENST00000592223.1 ENST00000329101.4 ENST00000586434.1 |
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr17_-_67138015 | 2.01 |
ENST00000284425.2
ENST00000590645.1 |
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr9_-_39288092 | 1.99 |
ENST00000323947.7
ENST00000297668.6 ENST00000377656.2 ENST00000377659.1 |
CNTNAP3
|
contactin associated protein-like 3 |
| chr4_-_70626430 | 1.86 |
ENST00000310613.3
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr15_-_52970820 | 1.84 |
ENST00000261844.7
ENST00000399202.4 ENST00000562135.1 |
FAM214A
|
family with sequence similarity 214, member A |
| chr2_-_165424973 | 1.78 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr4_-_102268628 | 1.62 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr12_+_96588279 | 1.61 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr7_+_12250886 | 1.53 |
ENST00000444443.1
ENST00000396667.3 |
TMEM106B
|
transmembrane protein 106B |
| chr18_-_53303123 | 1.44 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr7_+_134832808 | 1.40 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr6_+_123110465 | 1.39 |
ENST00000539041.1
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr1_+_87797351 | 1.38 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr3_+_132316081 | 1.35 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr2_+_109204743 | 1.35 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr4_-_90757364 | 1.31 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr4_-_90756769 | 1.31 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr9_-_86432547 | 1.23 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr18_+_6834472 | 1.19 |
ENST00000581099.1
ENST00000419673.2 ENST00000531294.1 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr11_-_89223883 | 1.13 |
ENST00000528341.1
|
NOX4
|
NADPH oxidase 4 |
| chr7_-_27702455 | 1.13 |
ENST00000265395.2
|
HIBADH
|
3-hydroxyisobutyrate dehydrogenase |
| chr10_-_25241499 | 1.13 |
ENST00000376378.1
ENST00000376376.3 ENST00000320152.6 |
PRTFDC1
|
phosphoribosyl transferase domain containing 1 |
| chr2_+_65283506 | 1.10 |
ENST00000377990.2
|
CEP68
|
centrosomal protein 68kDa |
| chr8_+_74903580 | 1.09 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr11_-_89224139 | 1.06 |
ENST00000413594.2
|
NOX4
|
NADPH oxidase 4 |
| chr2_+_109204909 | 1.06 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr15_-_31283798 | 1.05 |
ENST00000435680.1
ENST00000425768.1 |
MTMR10
|
myotubularin related protein 10 |
| chr3_-_28390581 | 1.05 |
ENST00000479665.1
|
AZI2
|
5-azacytidine induced 2 |
| chr3_+_124223586 | 1.05 |
ENST00000393496.1
|
KALRN
|
kalirin, RhoGEF kinase |
| chr9_+_27109133 | 1.04 |
ENST00000519097.1
ENST00000380036.4 |
TEK
|
TEK tyrosine kinase, endothelial |
| chr16_-_15736881 | 1.04 |
ENST00000540441.2
|
KIAA0430
|
KIAA0430 |
| chr3_-_141868293 | 1.03 |
ENST00000317104.7
ENST00000494358.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr17_-_67057047 | 1.02 |
ENST00000495634.1
ENST00000453985.2 ENST00000585714.1 |
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr9_-_13175823 | 1.02 |
ENST00000545857.1
|
MPDZ
|
multiple PDZ domain protein |
| chr3_-_101232019 | 0.99 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr8_-_6420565 | 0.99 |
ENST00000338312.6
|
ANGPT2
|
angiopoietin 2 |
| chr17_-_67057114 | 0.98 |
ENST00000370732.2
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr1_+_63989004 | 0.98 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr11_-_89224299 | 0.98 |
ENST00000343727.5
ENST00000531342.1 ENST00000375979.3 |
NOX4
|
NADPH oxidase 4 |
| chr4_-_152149033 | 0.97 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr1_+_145524891 | 0.95 |
ENST00000369304.3
|
ITGA10
|
integrin, alpha 10 |
| chr9_+_108006880 | 0.94 |
ENST00000374723.1
ENST00000374720.3 ENST00000374724.1 |
SLC44A1
|
solute carrier family 44 (choline transporter), member 1 |
| chr2_+_65283529 | 0.93 |
ENST00000546106.1
ENST00000537589.1 ENST00000260569.4 |
CEP68
|
centrosomal protein 68kDa |
| chr6_-_42016385 | 0.93 |
ENST00000502771.1
ENST00000508143.1 ENST00000514588.1 ENST00000510503.1 ENST00000415497.2 ENST00000372988.4 |
CCND3
|
cyclin D3 |
| chr19_+_18111927 | 0.92 |
ENST00000379656.3
|
ARRDC2
|
arrestin domain containing 2 |
| chrX_-_20236970 | 0.90 |
ENST00000379548.4
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr12_+_96588143 | 0.87 |
ENST00000228741.3
ENST00000547249.1 |
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr1_+_97188188 | 0.87 |
ENST00000541987.1
|
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr3_-_28390298 | 0.87 |
ENST00000457172.1
|
AZI2
|
5-azacytidine induced 2 |
| chr14_+_102276132 | 0.86 |
ENST00000350249.3
ENST00000557621.1 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr8_-_6420759 | 0.86 |
ENST00000523120.1
|
ANGPT2
|
angiopoietin 2 |
| chr2_-_101767715 | 0.85 |
ENST00000376840.4
ENST00000409318.1 |
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain) |
| chr12_-_71031185 | 0.84 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr14_-_103987679 | 0.84 |
ENST00000553610.1
|
CKB
|
creatine kinase, brain |
| chr3_+_122513642 | 0.84 |
ENST00000261038.5
|
DIRC2
|
disrupted in renal carcinoma 2 |
| chr13_+_103459704 | 0.80 |
ENST00000602836.1
|
BIVM-ERCC5
|
BIVM-ERCC5 readthrough |
| chr12_-_68696652 | 0.79 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr10_-_49482907 | 0.79 |
ENST00000374201.3
ENST00000407470.4 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr12_-_12491608 | 0.79 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr3_-_28390415 | 0.78 |
ENST00000414162.1
ENST00000420543.2 |
AZI2
|
5-azacytidine induced 2 |
| chr1_-_16763685 | 0.76 |
ENST00000540400.1
|
SPATA21
|
spermatogenesis associated 21 |
| chr13_-_41240717 | 0.76 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr2_+_66662510 | 0.75 |
ENST00000272369.9
ENST00000407092.2 |
MEIS1
|
Meis homeobox 1 |
| chr18_+_47088401 | 0.75 |
ENST00000261292.4
ENST00000427224.2 ENST00000580036.1 |
LIPG
|
lipase, endothelial |
| chr14_+_102276209 | 0.74 |
ENST00000445439.3
ENST00000334743.5 ENST00000557095.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr1_+_6845497 | 0.73 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr7_+_7811992 | 0.72 |
ENST00000406829.1
|
RPA3-AS1
|
RPA3 antisense RNA 1 |
| chr12_-_71031220 | 0.72 |
ENST00000334414.6
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr8_-_6420777 | 0.71 |
ENST00000415216.1
|
ANGPT2
|
angiopoietin 2 |
| chr11_+_844067 | 0.71 |
ENST00000397406.1
ENST00000409543.2 ENST00000525201.1 |
TSPAN4
|
tetraspanin 4 |
| chr5_-_42812143 | 0.71 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr13_-_99630233 | 0.70 |
ENST00000376460.1
ENST00000442173.1 |
DOCK9
|
dedicator of cytokinesis 9 |
| chr3_-_141868357 | 0.70 |
ENST00000489671.1
ENST00000475734.1 ENST00000467072.1 ENST00000499676.2 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr5_-_42811986 | 0.70 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr8_-_6420930 | 0.69 |
ENST00000325203.5
|
ANGPT2
|
angiopoietin 2 |
| chr1_-_234667504 | 0.69 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr5_-_10308125 | 0.66 |
ENST00000296658.3
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr2_+_191513959 | 0.66 |
ENST00000337386.5
ENST00000357215.5 |
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr18_-_53069419 | 0.65 |
ENST00000570177.2
|
TCF4
|
transcription factor 4 |
| chr11_-_94227029 | 0.62 |
ENST00000323977.3
ENST00000536754.1 ENST00000323929.3 |
MRE11A
|
MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
| chr7_+_77469439 | 0.62 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr4_+_40198527 | 0.61 |
ENST00000381799.5
|
RHOH
|
ras homolog family member H |
| chr11_+_844406 | 0.60 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr15_-_55657428 | 0.60 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr3_-_28390120 | 0.59 |
ENST00000334100.6
|
AZI2
|
5-azacytidine induced 2 |
| chrX_-_101112549 | 0.59 |
ENST00000537026.1
ENST00000361708.2 |
NXF5
|
nuclear RNA export factor 5 |
| chr7_-_95064264 | 0.58 |
ENST00000536183.1
ENST00000433091.2 ENST00000222572.3 |
PON2
|
paraoxonase 2 |
| chr14_+_50234827 | 0.58 |
ENST00000554589.1
ENST00000557247.1 |
KLHDC2
|
kelch domain containing 2 |
| chr16_+_28858004 | 0.58 |
ENST00000322610.8
|
SH2B1
|
SH2B adaptor protein 1 |
| chr12_-_102591604 | 0.58 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr18_-_30716038 | 0.57 |
ENST00000581852.1
|
CCDC178
|
coiled-coil domain containing 178 |
| chr6_-_167571817 | 0.55 |
ENST00000366834.1
|
GPR31
|
G protein-coupled receptor 31 |
| chr19_+_30863271 | 0.55 |
ENST00000355537.3
|
ZNF536
|
zinc finger protein 536 |
| chr12_-_772901 | 0.53 |
ENST00000305108.4
|
NINJ2
|
ninjurin 2 |
| chr6_-_13621126 | 0.52 |
ENST00000600057.1
|
AL441883.1
|
Uncharacterized protein |
| chr8_+_93895865 | 0.52 |
ENST00000391681.1
|
AC117834.1
|
AC117834.1 |
| chr18_-_21852143 | 0.52 |
ENST00000399443.3
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr9_-_88896977 | 0.51 |
ENST00000311534.6
|
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr6_-_31830655 | 0.51 |
ENST00000375631.4
|
NEU1
|
sialidase 1 (lysosomal sialidase) |
| chr17_+_67410832 | 0.51 |
ENST00000590474.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chrX_-_148676974 | 0.50 |
ENST00000524178.1
|
HSFX2
|
heat shock transcription factor family, X linked 2 |
| chr6_+_32121789 | 0.49 |
ENST00000437001.2
ENST00000375137.2 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr2_+_170683979 | 0.49 |
ENST00000418381.1
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr10_+_111985713 | 0.48 |
ENST00000239007.7
|
MXI1
|
MAX interactor 1, dimerization protein |
| chrX_+_148855726 | 0.48 |
ENST00000370416.4
|
HSFX1
|
heat shock transcription factor family, X linked 1 |
| chr6_+_32121908 | 0.47 |
ENST00000375143.2
ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr1_-_24469602 | 0.47 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1 |
| chr15_+_49462434 | 0.46 |
ENST00000558145.1
ENST00000543495.1 ENST00000544523.1 ENST00000560138.1 |
GALK2
|
galactokinase 2 |
| chr1_+_153940713 | 0.46 |
ENST00000368601.1
ENST00000368603.1 ENST00000368600.3 |
CREB3L4
|
cAMP responsive element binding protein 3-like 4 |
| chr1_-_54405773 | 0.46 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr6_-_105627735 | 0.46 |
ENST00000254765.3
|
POPDC3
|
popeye domain containing 3 |
| chr1_-_227505289 | 0.46 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr14_+_23067146 | 0.46 |
ENST00000428304.2
|
ABHD4
|
abhydrolase domain containing 4 |
| chr7_+_101460882 | 0.46 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr14_+_22985251 | 0.44 |
ENST00000390510.1
|
TRAJ27
|
T cell receptor alpha joining 27 |
| chr1_+_153940346 | 0.44 |
ENST00000405694.3
ENST00000449724.1 ENST00000368607.3 ENST00000271889.4 |
CREB3L4
|
cAMP responsive element binding protein 3-like 4 |
| chr3_+_159570722 | 0.44 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr7_-_6098770 | 0.43 |
ENST00000536084.1
ENST00000446699.1 ENST00000199389.6 |
EIF2AK1
|
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr2_+_170683942 | 0.43 |
ENST00000272793.5
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr11_-_82745238 | 0.43 |
ENST00000531021.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr19_-_40732594 | 0.42 |
ENST00000430325.2
ENST00000433940.1 |
CNTD2
|
cyclin N-terminal domain containing 2 |
| chr7_-_105332084 | 0.42 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr1_+_95616933 | 0.41 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr6_-_46889694 | 0.41 |
ENST00000283296.7
ENST00000362015.4 ENST00000456426.2 |
GPR116
|
G protein-coupled receptor 116 |
| chr12_+_118454500 | 0.40 |
ENST00000537315.1
ENST00000229043.3 ENST00000484086.2 ENST00000420967.1 ENST00000454402.2 ENST00000392542.2 ENST00000535092.1 |
RFC5
|
replication factor C (activator 1) 5, 36.5kDa |
| chr2_+_175260451 | 0.40 |
ENST00000458563.1
ENST00000409673.3 ENST00000272732.6 ENST00000435964.1 |
SCRN3
|
secernin 3 |
| chr11_-_62476965 | 0.39 |
ENST00000405837.1
ENST00000531524.1 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr11_-_62477041 | 0.39 |
ENST00000433053.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr3_+_107096188 | 0.39 |
ENST00000261058.1
|
CCDC54
|
coiled-coil domain containing 54 |
| chr3_-_157221128 | 0.39 |
ENST00000392833.2
ENST00000362010.2 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr4_-_140223670 | 0.38 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr5_-_107703556 | 0.38 |
ENST00000496714.1
|
FBXL17
|
F-box and leucine-rich repeat protein 17 |
| chr11_-_114466477 | 0.38 |
ENST00000375478.3
|
NXPE4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr7_-_38394118 | 0.38 |
ENST00000390345.2
|
TRGV4
|
T cell receptor gamma variable 4 |
| chr9_+_70856397 | 0.37 |
ENST00000360171.6
|
CBWD3
|
COBW domain containing 3 |
| chr1_+_117297007 | 0.36 |
ENST00000369478.3
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chr5_+_79703823 | 0.36 |
ENST00000338008.5
ENST00000510158.1 ENST00000505560.1 |
ZFYVE16
|
zinc finger, FYVE domain containing 16 |
| chr19_-_37019136 | 0.36 |
ENST00000592282.1
|
ZNF260
|
zinc finger protein 260 |
| chr5_-_122758994 | 0.36 |
ENST00000306467.5
ENST00000515110.1 |
CEP120
|
centrosomal protein 120kDa |
| chr12_-_109219937 | 0.36 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chr3_+_28390637 | 0.35 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr11_-_107328527 | 0.35 |
ENST00000282251.5
ENST00000433523.1 |
CWF19L2
|
CWF19-like 2, cell cycle control (S. pombe) |
| chr13_-_41837620 | 0.35 |
ENST00000379477.1
ENST00000452359.1 ENST00000379480.4 ENST00000430347.2 |
MTRF1
|
mitochondrial translational release factor 1 |
| chr1_-_241803649 | 0.35 |
ENST00000366554.2
|
OPN3
|
opsin 3 |
| chr19_+_57640011 | 0.34 |
ENST00000598197.1
|
USP29
|
ubiquitin specific peptidase 29 |
| chr5_-_125930929 | 0.34 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr2_-_192711968 | 0.34 |
ENST00000304141.4
|
SDPR
|
serum deprivation response |
| chr8_+_70404996 | 0.33 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr2_+_86947296 | 0.33 |
ENST00000283632.4
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| chr12_+_100041527 | 0.33 |
ENST00000324341.1
|
FAM71C
|
family with sequence similarity 71, member C |
| chr20_+_45338126 | 0.33 |
ENST00000359271.2
|
SLC2A10
|
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr1_-_182360918 | 0.33 |
ENST00000339526.4
|
GLUL
|
glutamate-ammonia ligase |
| chr1_+_214163033 | 0.33 |
ENST00000607425.1
|
PROX1
|
prospero homeobox 1 |
| chr9_+_42671887 | 0.32 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr20_+_18125727 | 0.32 |
ENST00000489634.2
|
CSRP2BP
|
CSRP2 binding protein |
| chr4_-_140223614 | 0.32 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr5_-_122759032 | 0.32 |
ENST00000510582.3
ENST00000328236.5 ENST00000306481.6 ENST00000508442.2 ENST00000395431.2 |
CEP120
|
centrosomal protein 120kDa |
| chr19_-_36523709 | 0.31 |
ENST00000592017.1
ENST00000360535.4 |
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr6_+_97010424 | 0.31 |
ENST00000541107.1
ENST00000450218.1 ENST00000326771.2 |
FHL5
|
four and a half LIM domains 5 |
| chr10_-_11574274 | 0.31 |
ENST00000277575.5
|
USP6NL
|
USP6 N-terminal like |
| chr17_-_39150385 | 0.31 |
ENST00000391586.1
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr1_+_28844778 | 0.31 |
ENST00000411533.1
|
RCC1
|
regulator of chromosome condensation 1 |
| chr1_-_241803679 | 0.31 |
ENST00000331838.5
|
OPN3
|
opsin 3 |
| chr9_+_706842 | 0.31 |
ENST00000382293.3
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr8_-_62602327 | 0.30 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr1_+_151739131 | 0.30 |
ENST00000400999.1
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr1_-_47655686 | 0.30 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr11_+_128634589 | 0.30 |
ENST00000281428.8
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chrX_-_19689106 | 0.29 |
ENST00000379716.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr14_+_70918874 | 0.29 |
ENST00000603540.1
|
ADAM21
|
ADAM metallopeptidase domain 21 |
| chr3_+_101292939 | 0.29 |
ENST00000265260.3
ENST00000469941.1 ENST00000296024.5 |
PCNP
|
PEST proteolytic signal containing nuclear protein |
| chr11_+_12115543 | 0.29 |
ENST00000537344.1
ENST00000532179.1 ENST00000526065.1 |
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr4_+_118955500 | 0.29 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr8_-_133772794 | 0.28 |
ENST00000519187.1
ENST00000523829.1 ENST00000356838.3 ENST00000377901.4 ENST00000519304.1 |
TMEM71
|
transmembrane protein 71 |
| chr14_-_92572894 | 0.28 |
ENST00000532032.1
ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3
|
ataxin 3 |
| chr1_-_168106536 | 0.28 |
ENST00000537209.1
ENST00000361697.2 ENST00000546300.1 ENST00000367835.1 |
GPR161
|
G protein-coupled receptor 161 |
| chr11_-_118095718 | 0.28 |
ENST00000526620.1
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1 |
| chr1_-_182360498 | 0.28 |
ENST00000417584.2
|
GLUL
|
glutamate-ammonia ligase |
| chr12_-_102872317 | 0.28 |
ENST00000424202.2
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr17_+_3379284 | 0.28 |
ENST00000263080.2
|
ASPA
|
aspartoacylase |
| chr8_-_54755789 | 0.28 |
ENST00000359530.2
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr1_-_16400086 | 0.28 |
ENST00000375662.4
|
FAM131C
|
family with sequence similarity 131, member C |
| chr19_-_7293942 | 0.27 |
ENST00000341500.5
ENST00000302850.5 |
INSR
|
insulin receptor |
| chr3_+_48956249 | 0.27 |
ENST00000452882.1
ENST00000430423.1 ENST00000356401.4 ENST00000449376.1 ENST00000420814.1 ENST00000449729.1 ENST00000433170.1 |
ARIH2
|
ariadne RBR E3 ubiquitin protein ligase 2 |
| chr12_-_24103954 | 0.27 |
ENST00000441133.2
ENST00000545921.1 |
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr16_+_14980632 | 0.27 |
ENST00000565655.1
|
NOMO1
|
NODAL modulator 1 |
| chr19_+_17378278 | 0.27 |
ENST00000596335.1
ENST00000601436.1 ENST00000595632.1 |
BABAM1
|
BRISC and BRCA1 A complex member 1 |
| chr8_+_21777159 | 0.26 |
ENST00000434536.1
ENST00000252512.9 |
XPO7
|
exportin 7 |
| chr4_-_14889791 | 0.26 |
ENST00000509654.1
ENST00000515031.1 ENST00000505089.2 |
LINC00504
|
long intergenic non-protein coding RNA 504 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXO4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 1.1 | 7.6 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.7 | 3.3 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.5 | 1.6 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.5 | 2.6 | GO:1903285 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.4 | 2.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.4 | 1.2 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.4 | 1.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.4 | 2.9 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.3 | 9.5 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.3 | 0.5 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.2 | 0.8 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.2 | 1.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.2 | 0.7 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.2 | 0.9 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 | 4.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.2 | 1.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.2 | 0.6 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 0.8 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.1 | 1.0 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 1.3 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.1 | 2.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.6 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.4 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.1 | 2.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.3 | GO:0090425 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.1 | 0.8 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.4 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.1 | 1.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.3 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.3 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 1.0 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.3 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.5 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.3 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.1 | 1.4 | GO:0042659 | spinal cord association neuron differentiation(GO:0021527) regulation of cell fate specification(GO:0042659) |
| 0.1 | 3.2 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 0.7 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 1.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.3 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.3 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 0.3 | GO:1990535 | transformation of host cell by virus(GO:0019087) neuron projection maintenance(GO:1990535) |
| 0.1 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 | 0.2 | GO:0002840 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.1 | 0.8 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.4 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.2 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.1 | 2.4 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 | 0.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.5 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.2 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 0.4 | GO:1902715 | regulation of myeloid dendritic cell activation(GO:0030885) positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.1 | GO:0035565 | regulation of pronephros size(GO:0035565) renal glucose absorption(GO:0035623) |
| 0.0 | 1.1 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 1.8 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 1.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.2 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.0 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.8 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.9 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.2 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.2 | GO:0046606 | negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.5 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.6 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.8 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.5 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 1.3 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.9 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 0.2 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 1.6 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.5 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.0 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.5 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.5 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.7 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 1.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.2 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.3 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.0 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.0 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.5 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.2 | 1.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 1.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 3.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.3 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 2.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 3.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 2.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 1.0 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 0.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.3 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 1.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 8.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.6 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 1.7 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 2.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 2.1 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 2.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0097342 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 0.7 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.5 | 2.6 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.3 | 9.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.3 | 1.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.3 | 5.4 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.2 | 3.2 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.2 | 2.0 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.2 | 0.6 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.2 | 4.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 1.6 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 2.1 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 1.0 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 0.5 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.2 | 0.6 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 1.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.4 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 1.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.9 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.3 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 1.0 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.7 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.5 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.1 | 0.5 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.5 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.2 | GO:0004917 | interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.6 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.3 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 1.6 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.4 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.3 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.3 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 1.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.8 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 1.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 1.1 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 1.7 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.5 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.6 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 4.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.9 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.8 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 1.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 9.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 3.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.9 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.8 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 2.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 2.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 4.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 4.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 2.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.9 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.7 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 2.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.8 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 2.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 2.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |