Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
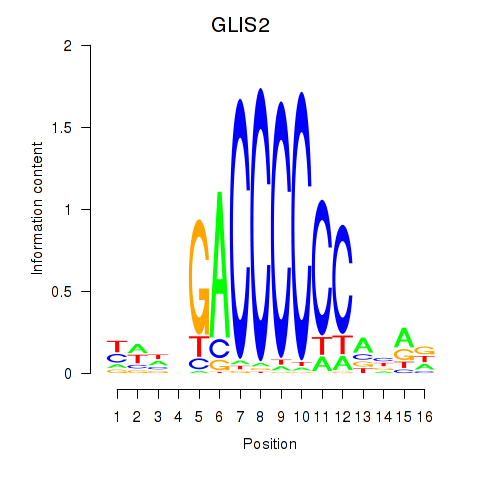
Results for GLIS2
Z-value: 0.48
Transcription factors associated with GLIS2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLIS2
|
ENSG00000126603.4 | GLIS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLIS2 | hg19_v2_chr16_+_4364762_4364762, hg19_v2_chr16_+_4382225_4382225 | -0.31 | 1.3e-01 | Click! |
Activity profile of GLIS2 motif
Sorted Z-values of GLIS2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_45973120 | 2.07 |
ENST00000592811.1
ENST00000586615.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr3_-_18466026 | 1.44 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr5_+_68788594 | 1.37 |
ENST00000396442.2
ENST00000380766.2 |
OCLN
|
occludin |
| chr2_+_48757278 | 1.25 |
ENST00000404752.1
ENST00000406226.1 |
STON1
|
stonin 1 |
| chr1_-_92371839 | 0.97 |
ENST00000370399.2
|
TGFBR3
|
transforming growth factor, beta receptor III |
| chr22_+_31523734 | 0.80 |
ENST00000402238.1
ENST00000404453.1 ENST00000401755.1 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr14_-_105635090 | 0.80 |
ENST00000331782.3
ENST00000347004.2 |
JAG2
|
jagged 2 |
| chr1_-_186649543 | 0.65 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr16_+_55542910 | 0.64 |
ENST00000262134.5
|
LPCAT2
|
lysophosphatidylcholine acyltransferase 2 |
| chr6_+_84743436 | 0.59 |
ENST00000257776.4
|
MRAP2
|
melanocortin 2 receptor accessory protein 2 |
| chr14_-_54955721 | 0.57 |
ENST00000554908.1
|
GMFB
|
glia maturation factor, beta |
| chr10_+_94608218 | 0.55 |
ENST00000371543.1
|
EXOC6
|
exocyst complex component 6 |
| chr5_+_140566 | 0.54 |
ENST00000502646.1
|
PLEKHG4B
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4B |
| chr1_-_20503917 | 0.52 |
ENST00000429261.2
|
PLA2G2C
|
phospholipase A2, group IIC |
| chr20_+_30598231 | 0.52 |
ENST00000300415.8
ENST00000262659.8 |
CCM2L
|
cerebral cavernous malformation 2-like |
| chrX_+_46433193 | 0.45 |
ENST00000276055.3
|
CHST7
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
| chr19_+_11200038 | 0.45 |
ENST00000558518.1
ENST00000557933.1 ENST00000455727.2 ENST00000535915.1 ENST00000545707.1 ENST00000558013.1 |
LDLR
|
low density lipoprotein receptor |
| chr22_-_28197486 | 0.45 |
ENST00000302326.4
|
MN1
|
meningioma (disrupted in balanced translocation) 1 |
| chr1_-_24127256 | 0.43 |
ENST00000418277.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr9_+_139839711 | 0.42 |
ENST00000224181.3
|
C8G
|
complement component 8, gamma polypeptide |
| chr2_-_113999260 | 0.41 |
ENST00000468980.2
|
PAX8
|
paired box 8 |
| chrX_+_118108571 | 0.40 |
ENST00000304778.7
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr19_+_50979753 | 0.39 |
ENST00000597426.1
ENST00000334976.6 ENST00000376918.3 ENST00000598585.1 |
EMC10
|
ER membrane protein complex subunit 10 |
| chr17_+_61571746 | 0.39 |
ENST00000579409.1
|
ACE
|
angiotensin I converting enzyme |
| chr21_-_31869451 | 0.39 |
ENST00000334058.2
|
KRTAP19-4
|
keratin associated protein 19-4 |
| chr6_-_2903514 | 0.39 |
ENST00000380698.4
|
SERPINB9
|
serpin peptidase inhibitor, clade B (ovalbumin), member 9 |
| chr2_+_24272576 | 0.38 |
ENST00000380986.4
ENST00000452109.1 |
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa |
| chr3_-_125802765 | 0.38 |
ENST00000514891.1
ENST00000512470.1 ENST00000504035.1 ENST00000360370.4 ENST00000513723.1 ENST00000510651.1 ENST00000514333.1 |
SLC41A3
|
solute carrier family 41, member 3 |
| chr11_+_86748863 | 0.37 |
ENST00000340353.7
|
TMEM135
|
transmembrane protein 135 |
| chr1_+_44412577 | 0.37 |
ENST00000372343.3
|
IPO13
|
importin 13 |
| chr10_-_33623564 | 0.37 |
ENST00000374875.1
ENST00000374822.4 |
NRP1
|
neuropilin 1 |
| chr19_-_2050852 | 0.37 |
ENST00000541165.1
ENST00000591601.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr9_-_34710066 | 0.37 |
ENST00000378792.1
ENST00000259607.2 |
CCL21
|
chemokine (C-C motif) ligand 21 |
| chr6_-_133119668 | 0.34 |
ENST00000275227.4
ENST00000538764.1 |
SLC18B1
|
solute carrier family 18, subfamily B, member 1 |
| chr11_+_125034586 | 0.34 |
ENST00000298282.9
|
PKNOX2
|
PBX/knotted 1 homeobox 2 |
| chrX_-_70474910 | 0.33 |
ENST00000373988.1
ENST00000373998.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr19_-_41859814 | 0.33 |
ENST00000221930.5
|
TGFB1
|
transforming growth factor, beta 1 |
| chr3_+_44626446 | 0.33 |
ENST00000441021.1
ENST00000322734.2 |
ZNF660
|
zinc finger protein 660 |
| chr11_-_2160180 | 0.33 |
ENST00000381406.4
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chrX_+_118108601 | 0.32 |
ENST00000371628.3
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr8_+_118533049 | 0.32 |
ENST00000522839.1
|
MED30
|
mediator complex subunit 30 |
| chr10_+_94608245 | 0.31 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr11_+_844067 | 0.31 |
ENST00000397406.1
ENST00000409543.2 ENST00000525201.1 |
TSPAN4
|
tetraspanin 4 |
| chr20_-_30310797 | 0.31 |
ENST00000422920.1
|
BCL2L1
|
BCL2-like 1 |
| chr3_-_125803105 | 0.31 |
ENST00000346785.5
ENST00000315891.6 |
SLC41A3
|
solute carrier family 41, member 3 |
| chr4_-_107237340 | 0.31 |
ENST00000394706.3
|
TBCK
|
TBC1 domain containing kinase |
| chr8_-_29120580 | 0.31 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr4_-_107237374 | 0.30 |
ENST00000361687.4
ENST00000507696.1 ENST00000394708.2 ENST00000509532.1 |
TBCK
|
TBC1 domain containing kinase |
| chr19_+_49891475 | 0.30 |
ENST00000447857.3
|
CCDC155
|
coiled-coil domain containing 155 |
| chr20_-_30458019 | 0.30 |
ENST00000486996.1
ENST00000398084.2 |
DUSP15
|
dual specificity phosphatase 15 |
| chr6_-_119670919 | 0.28 |
ENST00000368468.3
|
MAN1A1
|
mannosidase, alpha, class 1A, member 1 |
| chr17_-_5138099 | 0.28 |
ENST00000571800.1
ENST00000574081.1 ENST00000399600.4 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chr9_-_35112376 | 0.28 |
ENST00000488109.2
|
FAM214B
|
family with sequence similarity 214, member B |
| chr10_-_75634219 | 0.28 |
ENST00000305762.7
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr2_+_24272543 | 0.27 |
ENST00000380991.4
|
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa |
| chr22_-_38794490 | 0.27 |
ENST00000400206.2
|
CSNK1E
|
casein kinase 1, epsilon |
| chr15_-_31283798 | 0.27 |
ENST00000435680.1
ENST00000425768.1 |
MTMR10
|
myotubularin related protein 10 |
| chr3_-_197282821 | 0.27 |
ENST00000445160.2
ENST00000446746.1 ENST00000432819.1 ENST00000392379.1 ENST00000441275.1 ENST00000392378.2 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr15_-_90233866 | 0.27 |
ENST00000561257.1
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr22_-_21482046 | 0.26 |
ENST00000419447.1
|
POM121L7
|
POM121 transmembrane nucleoporin-like 7 |
| chr11_+_31531291 | 0.26 |
ENST00000350638.5
ENST00000379163.5 ENST00000395934.2 |
ELP4
|
elongator acetyltransferase complex subunit 4 |
| chr22_-_50700140 | 0.26 |
ENST00000215659.8
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr19_+_10527449 | 0.26 |
ENST00000592685.1
ENST00000380702.2 |
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr14_+_23842018 | 0.26 |
ENST00000397242.2
ENST00000329715.2 |
IL25
|
interleukin 25 |
| chr16_-_4664860 | 0.26 |
ENST00000587615.1
ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1
|
UBA-like domain containing 1 |
| chr9_+_134103496 | 0.25 |
ENST00000498010.1
ENST00000476004.1 ENST00000528406.1 |
NUP214
|
nucleoporin 214kDa |
| chr20_-_30310656 | 0.25 |
ENST00000376055.4
|
BCL2L1
|
BCL2-like 1 |
| chr14_-_105531759 | 0.24 |
ENST00000329797.3
ENST00000539291.2 ENST00000392585.2 |
GPR132
|
G protein-coupled receptor 132 |
| chr11_-_31531121 | 0.24 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr19_-_42759300 | 0.24 |
ENST00000222329.4
|
ERF
|
Ets2 repressor factor |
| chrX_-_10557949 | 0.23 |
ENST00000380780.1
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr12_+_57853918 | 0.23 |
ENST00000532291.1
ENST00000543426.1 ENST00000228682.2 ENST00000546141.1 |
GLI1
|
GLI family zinc finger 1 |
| chr19_+_36602104 | 0.23 |
ENST00000585332.1
ENST00000262637.4 |
OVOL3
|
ovo-like zinc finger 3 |
| chr16_-_3306587 | 0.23 |
ENST00000541159.1
ENST00000536379.1 ENST00000219596.1 ENST00000339854.4 |
MEFV
|
Mediterranean fever |
| chr19_-_17185848 | 0.23 |
ENST00000593360.1
|
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr19_+_45409011 | 0.22 |
ENST00000252486.4
ENST00000446996.1 ENST00000434152.1 |
APOE
|
apolipoprotein E |
| chr20_-_30310693 | 0.22 |
ENST00000307677.4
ENST00000420653.1 |
BCL2L1
|
BCL2-like 1 |
| chr10_+_14880287 | 0.21 |
ENST00000437161.2
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr16_-_49890016 | 0.21 |
ENST00000563137.2
|
ZNF423
|
zinc finger protein 423 |
| chr19_+_1905365 | 0.20 |
ENST00000329478.2
ENST00000602400.1 ENST00000409472.1 |
ADAT3
SCAMP4
|
adenosine deaminase, tRNA-specific 3 secretory carrier membrane protein 4 |
| chr2_+_181845074 | 0.20 |
ENST00000602959.1
ENST00000602479.1 ENST00000392415.2 ENST00000602291.1 |
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr1_+_64936428 | 0.20 |
ENST00000371073.2
ENST00000290039.5 |
CACHD1
|
cache domain containing 1 |
| chr1_-_54411255 | 0.20 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr14_-_23426270 | 0.20 |
ENST00000557591.1
ENST00000397409.4 ENST00000490506.1 ENST00000554406.1 |
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr12_+_57998400 | 0.20 |
ENST00000548804.1
ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr20_+_33462888 | 0.20 |
ENST00000336325.4
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2 |
| chr14_-_23426322 | 0.19 |
ENST00000555367.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr14_-_23426337 | 0.19 |
ENST00000342454.8
ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr1_+_150980889 | 0.19 |
ENST00000450884.1
ENST00000271620.3 ENST00000271619.8 ENST00000368937.1 ENST00000431193.1 ENST00000368936.1 |
PRUNE
|
prune exopolyphosphatase |
| chr2_+_131113609 | 0.19 |
ENST00000347849.3
|
PTPN18
|
protein tyrosine phosphatase, non-receptor type 18 (brain-derived) |
| chr11_-_76381029 | 0.19 |
ENST00000407242.2
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr11_+_66624527 | 0.19 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr22_+_31489344 | 0.19 |
ENST00000404574.1
|
SMTN
|
smoothelin |
| chr11_-_1036706 | 0.19 |
ENST00000421673.2
|
MUC6
|
mucin 6, oligomeric mucus/gel-forming |
| chr16_-_67185117 | 0.19 |
ENST00000449549.3
|
B3GNT9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr7_+_133812052 | 0.18 |
ENST00000285928.2
|
LRGUK
|
leucine-rich repeats and guanylate kinase domain containing |
| chr9_-_98279241 | 0.18 |
ENST00000437951.1
ENST00000375274.2 ENST00000430669.2 ENST00000468211.2 |
PTCH1
|
patched 1 |
| chr19_-_46405861 | 0.18 |
ENST00000322217.5
|
MYPOP
|
Myb-related transcription factor, partner of profilin |
| chr19_-_19006920 | 0.18 |
ENST00000429504.2
ENST00000427170.2 |
CERS1
|
ceramide synthase 1 |
| chr4_-_106394866 | 0.18 |
ENST00000502596.1
|
PPA2
|
pyrophosphatase (inorganic) 2 |
| chr17_-_76921459 | 0.18 |
ENST00000262768.7
|
TIMP2
|
TIMP metallopeptidase inhibitor 2 |
| chr15_+_85359911 | 0.18 |
ENST00000258888.5
|
ALPK3
|
alpha-kinase 3 |
| chr17_+_78075361 | 0.18 |
ENST00000577106.1
ENST00000390015.3 |
GAA
|
glucosidase, alpha; acid |
| chr15_-_90234006 | 0.17 |
ENST00000300056.3
ENST00000559170.1 |
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr8_+_1772132 | 0.17 |
ENST00000349830.3
ENST00000520359.1 ENST00000518288.1 ENST00000398560.1 |
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr11_+_121447469 | 0.17 |
ENST00000532694.1
ENST00000534286.1 |
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr16_-_28518153 | 0.17 |
ENST00000356897.1
|
IL27
|
interleukin 27 |
| chr12_+_57998595 | 0.17 |
ENST00000337737.3
ENST00000548198.1 ENST00000551632.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr20_-_35492048 | 0.17 |
ENST00000237536.4
|
SOGA1
|
suppressor of glucose, autophagy associated 1 |
| chr2_+_169312350 | 0.17 |
ENST00000305747.6
|
CERS6
|
ceramide synthase 6 |
| chrX_+_152240819 | 0.17 |
ENST00000421798.3
ENST00000535416.1 |
PNMA6C
PNMA6A
|
paraneoplastic Ma antigen family member 6C paraneoplastic Ma antigen family member 6A |
| chrX_+_53449839 | 0.17 |
ENST00000457095.1
|
RIBC1
|
RIB43A domain with coiled-coils 1 |
| chr16_+_3070356 | 0.16 |
ENST00000341627.5
ENST00000575124.1 ENST00000575836.1 |
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr7_-_4998802 | 0.16 |
ENST00000406755.1
ENST00000404774.3 ENST00000401401.3 |
MMD2
|
monocyte to macrophage differentiation-associated 2 |
| chr7_+_140373619 | 0.16 |
ENST00000483369.1
|
ADCK2
|
aarF domain containing kinase 2 |
| chr16_+_3070313 | 0.16 |
ENST00000326577.4
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr12_-_133263893 | 0.16 |
ENST00000535270.1
ENST00000320574.5 |
POLE
|
polymerase (DNA directed), epsilon, catalytic subunit |
| chr2_+_131113580 | 0.16 |
ENST00000175756.5
|
PTPN18
|
protein tyrosine phosphatase, non-receptor type 18 (brain-derived) |
| chr1_-_201123546 | 0.16 |
ENST00000435310.1
ENST00000485839.2 ENST00000367330.1 |
TMEM9
|
transmembrane protein 9 |
| chr19_-_3061397 | 0.16 |
ENST00000586839.1
|
AES
|
amino-terminal enhancer of split |
| chr9_+_127539481 | 0.16 |
ENST00000373580.3
|
OLFML2A
|
olfactomedin-like 2A |
| chr11_+_66045634 | 0.16 |
ENST00000528852.1
ENST00000311445.6 |
CNIH2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr16_+_765092 | 0.16 |
ENST00000568223.2
|
METRN
|
meteorin, glial cell differentiation regulator |
| chr2_+_108905325 | 0.15 |
ENST00000438339.1
ENST00000409880.1 ENST00000437390.2 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr15_+_91478493 | 0.15 |
ENST00000418476.2
|
UNC45A
|
unc-45 homolog A (C. elegans) |
| chr19_+_11909329 | 0.15 |
ENST00000323169.5
ENST00000450087.1 |
ZNF491
|
zinc finger protein 491 |
| chr1_-_201123586 | 0.15 |
ENST00000414605.2
ENST00000367334.5 ENST00000367332.1 |
TMEM9
|
transmembrane protein 9 |
| chr1_+_201979645 | 0.15 |
ENST00000367284.5
ENST00000367283.3 |
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr6_-_39282221 | 0.15 |
ENST00000453413.2
|
KCNK17
|
potassium channel, subfamily K, member 17 |
| chr10_+_70320413 | 0.15 |
ENST00000373644.4
|
TET1
|
tet methylcytosine dioxygenase 1 |
| chr17_+_7788104 | 0.14 |
ENST00000380358.4
|
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr5_-_176836577 | 0.14 |
ENST00000253496.3
|
F12
|
coagulation factor XII (Hageman factor) |
| chr3_-_28390581 | 0.14 |
ENST00000479665.1
|
AZI2
|
5-azacytidine induced 2 |
| chr19_+_17337473 | 0.14 |
ENST00000598068.1
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr7_-_752577 | 0.14 |
ENST00000544935.1
ENST00000430040.1 ENST00000456696.2 ENST00000406797.1 |
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr1_-_32110467 | 0.14 |
ENST00000440872.2
ENST00000373703.4 |
PEF1
|
penta-EF-hand domain containing 1 |
| chr17_-_27224621 | 0.14 |
ENST00000394906.2
ENST00000585169.1 ENST00000394908.4 |
FLOT2
|
flotillin 2 |
| chr7_+_140372953 | 0.14 |
ENST00000072869.4
ENST00000476491.1 |
ADCK2
|
aarF domain containing kinase 2 |
| chr19_+_41103063 | 0.14 |
ENST00000308370.7
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr19_-_18653781 | 0.14 |
ENST00000596558.2
ENST00000453489.2 |
FKBP8
|
FK506 binding protein 8, 38kDa |
| chr7_-_130080977 | 0.14 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr21_-_45671014 | 0.13 |
ENST00000436357.1
|
DNMT3L
|
DNA (cytosine-5-)-methyltransferase 3-like |
| chrX_-_102319092 | 0.13 |
ENST00000372728.3
|
BEX1
|
brain expressed, X-linked 1 |
| chr21_+_42539701 | 0.13 |
ENST00000330333.6
ENST00000328735.6 ENST00000347667.5 |
BACE2
|
beta-site APP-cleaving enzyme 2 |
| chr19_+_14640372 | 0.13 |
ENST00000215567.5
ENST00000598298.1 ENST00000596073.1 ENST00000600083.1 ENST00000436007.2 |
TECR
|
trans-2,3-enoyl-CoA reductase |
| chr1_+_41157421 | 0.13 |
ENST00000372654.1
|
NFYC
|
nuclear transcription factor Y, gamma |
| chr11_-_119217365 | 0.13 |
ENST00000360167.4
ENST00000555262.1 ENST00000449574.2 ENST00000445041.2 |
MFRP
C1QTNF5
|
membrane frizzled-related protein C1q and tumor necrosis factor related protein 5 |
| chr6_-_39282329 | 0.13 |
ENST00000373231.4
|
KCNK17
|
potassium channel, subfamily K, member 17 |
| chr1_-_54411240 | 0.13 |
ENST00000371378.2
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr19_+_17337406 | 0.13 |
ENST00000597836.1
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr20_-_30433396 | 0.13 |
ENST00000375978.3
|
FOXS1
|
forkhead box S1 |
| chr5_+_139027877 | 0.13 |
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5 |
| chr19_+_56159509 | 0.13 |
ENST00000586790.1
ENST00000591578.1 ENST00000588740.1 |
CCDC106
|
coiled-coil domain containing 106 |
| chr3_-_48470838 | 0.13 |
ENST00000358459.4
ENST00000358536.4 |
PLXNB1
|
plexin B1 |
| chr7_+_2671663 | 0.13 |
ENST00000407643.1
|
TTYH3
|
tweety family member 3 |
| chr20_+_57226284 | 0.13 |
ENST00000458280.1
ENST00000355957.5 ENST00000361770.5 ENST00000312283.8 ENST00000412911.1 ENST00000359617.4 ENST00000371141.4 |
STX16
|
syntaxin 16 |
| chr19_+_55105085 | 0.13 |
ENST00000251372.3
ENST00000453777.1 |
LILRA1
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 1 |
| chr2_-_89310012 | 0.12 |
ENST00000493819.1
|
IGKV1-9
|
immunoglobulin kappa variable 1-9 |
| chr19_-_54850417 | 0.12 |
ENST00000291759.4
|
LILRA4
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 4 |
| chr6_-_32908765 | 0.12 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr2_-_179343268 | 0.12 |
ENST00000424785.2
|
FKBP7
|
FK506 binding protein 7 |
| chr20_+_57226841 | 0.12 |
ENST00000358029.4
ENST00000361830.3 |
STX16
|
syntaxin 16 |
| chr22_-_19974616 | 0.12 |
ENST00000344269.3
ENST00000401994.1 ENST00000406522.1 |
ARVCF
|
armadillo repeat gene deleted in velocardiofacial syndrome |
| chr3_-_28390298 | 0.12 |
ENST00000457172.1
|
AZI2
|
5-azacytidine induced 2 |
| chr2_-_85829496 | 0.12 |
ENST00000409668.1
|
TMEM150A
|
transmembrane protein 150A |
| chr1_-_40157345 | 0.12 |
ENST00000372844.3
|
HPCAL4
|
hippocalcin like 4 |
| chr1_+_150954493 | 0.12 |
ENST00000368947.4
|
ANXA9
|
annexin A9 |
| chrX_+_153656978 | 0.12 |
ENST00000369762.2
ENST00000422890.1 |
ATP6AP1
|
ATPase, H+ transporting, lysosomal accessory protein 1 |
| chr6_+_17393839 | 0.12 |
ENST00000489374.1
ENST00000378990.2 |
CAP2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr16_+_811073 | 0.11 |
ENST00000382862.3
ENST00000563651.1 |
MSLN
|
mesothelin |
| chr1_+_41157671 | 0.11 |
ENST00000534399.1
ENST00000372653.1 |
NFYC
|
nuclear transcription factor Y, gamma |
| chr19_-_41256207 | 0.11 |
ENST00000598485.2
ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr8_+_38243951 | 0.11 |
ENST00000297720.5
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr19_-_35454953 | 0.11 |
ENST00000404801.1
|
ZNF792
|
zinc finger protein 792 |
| chr6_+_33048222 | 0.11 |
ENST00000428835.1
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chrX_+_54834159 | 0.11 |
ENST00000375053.2
ENST00000347546.4 ENST00000375062.4 |
MAGED2
|
melanoma antigen family D, 2 |
| chr13_-_42535214 | 0.11 |
ENST00000379310.3
ENST00000281496.6 |
VWA8
|
von Willebrand factor A domain containing 8 |
| chr3_-_11888246 | 0.11 |
ENST00000455809.1
ENST00000444133.2 ENST00000273037.5 |
TAMM41
|
TAM41, mitochondrial translocator assembly and maintenance protein, homolog (S. cerevisiae) |
| chr11_-_64510409 | 0.11 |
ENST00000394429.1
ENST00000394428.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr19_-_2702681 | 0.11 |
ENST00000382159.3
|
GNG7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr20_-_52687059 | 0.11 |
ENST00000371435.2
ENST00000395961.3 |
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr12_+_50451331 | 0.11 |
ENST00000228468.4
|
ASIC1
|
acid-sensing (proton-gated) ion channel 1 |
| chr17_+_38278530 | 0.11 |
ENST00000398532.4
|
MSL1
|
male-specific lethal 1 homolog (Drosophila) |
| chr19_-_821931 | 0.11 |
ENST00000359894.2
ENST00000520876.3 ENST00000519502.1 |
LPPR3
|
hsa-mir-3187 |
| chr1_-_156675368 | 0.11 |
ENST00000368222.3
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr6_+_28109703 | 0.11 |
ENST00000457389.2
ENST00000330236.6 |
ZKSCAN8
|
zinc finger with KRAB and SCAN domains 8 |
| chr3_+_52017454 | 0.10 |
ENST00000476854.1
ENST00000476351.1 ENST00000494103.1 ENST00000404366.2 ENST00000469863.1 |
ACY1
|
aminoacylase 1 |
| chrX_+_53449805 | 0.10 |
ENST00000414955.2
|
RIBC1
|
RIB43A domain with coiled-coils 1 |
| chr10_-_104179682 | 0.10 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr1_-_242162375 | 0.10 |
ENST00000357246.3
|
MAP1LC3C
|
microtubule-associated protein 1 light chain 3 gamma |
| chr21_-_39288743 | 0.10 |
ENST00000609713.1
|
KCNJ6
|
potassium inwardly-rectifying channel, subfamily J, member 6 |
| chr5_-_1524015 | 0.10 |
ENST00000283415.3
|
LPCAT1
|
lysophosphatidylcholine acyltransferase 1 |
| chr19_+_56159362 | 0.10 |
ENST00000593069.1
ENST00000308964.3 |
CCDC106
|
coiled-coil domain containing 106 |
| chr19_-_4517613 | 0.10 |
ENST00000301286.3
|
PLIN4
|
perilipin 4 |
| chr11_-_796197 | 0.10 |
ENST00000530360.1
ENST00000528606.1 ENST00000320230.5 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr17_+_75315534 | 0.10 |
ENST00000590294.1
ENST00000329047.8 |
SEPT9
|
septin 9 |
| chr17_+_48503603 | 0.10 |
ENST00000502667.1
|
ACSF2
|
acyl-CoA synthetase family member 2 |
| chr2_-_208490027 | 0.10 |
ENST00000458426.1
ENST00000406927.2 ENST00000425132.1 |
METTL21A
|
methyltransferase like 21A |
| chr11_+_64058758 | 0.10 |
ENST00000538767.1
|
KCNK4
|
potassium channel, subfamily K, member 4 |
| chr17_+_73996987 | 0.10 |
ENST00000588812.1
ENST00000448471.1 |
CDK3
|
cyclin-dependent kinase 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GLIS2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.2 | 1.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 | 0.6 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.4 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 0.4 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.4 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.1 | 0.5 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 0.3 | GO:1900126 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 0.8 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.4 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.1 | 0.8 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 1.4 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.1 | 0.7 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.1 | 0.6 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 1.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 2.1 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.1 | 0.4 | GO:0021649 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.1 | 0.4 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.1 | 0.2 | GO:1902994 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 0.4 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.2 | GO:0036146 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.1 | 0.2 | GO:1902771 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.2 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.1 | 0.4 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.2 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.2 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.0 | 0.2 | GO:0019413 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.3 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.2 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.2 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.2 | GO:0021938 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.2 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.3 | GO:0009624 | response to nematode(GO:0009624) eosinophil differentiation(GO:0030222) |
| 0.0 | 0.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.3 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.2 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.8 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.3 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.1 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.5 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.1 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:1990736 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.0 | 0.1 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.3 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0090035 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.3 | GO:0090220 | chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.0 | 0.0 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.3 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.0 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.5 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.2 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.0 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.3 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 0.0 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.5 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.2 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.6 | GO:0097009 | energy homeostasis(GO:0097009) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 0.5 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.1 | 0.2 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.7 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 1.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.0 | 0.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 1.4 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.2 | 0.6 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.2 | 0.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.7 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 1.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.4 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.1 | 0.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.8 | GO:0034485 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 0.5 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.4 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 0.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.4 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.1 | 0.2 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.4 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.3 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.0 | GO:0097259 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.0 | 0.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.0 | GO:0032551 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.7 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 2.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 0.8 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.0 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |