Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
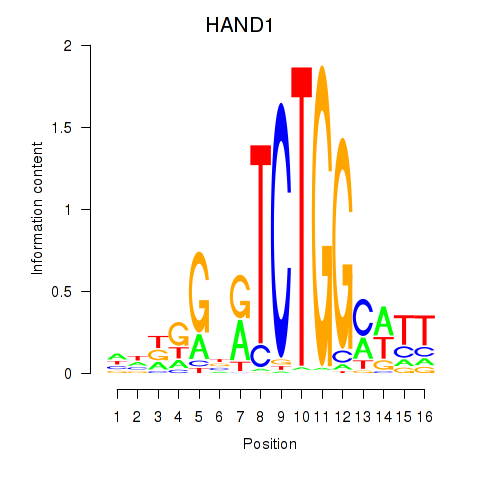
Results for HAND1
Z-value: 0.51
Transcription factors associated with HAND1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HAND1
|
ENSG00000113196.2 | heart and neural crest derivatives expressed 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HAND1 | hg19_v2_chr5_-_153857819_153857824 | -0.29 | 1.6e-01 | Click! |
Activity profile of HAND1 motif
Sorted Z-values of HAND1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_228678550 | 1.50 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr4_+_41614720 | 1.04 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr4_+_41614909 | 1.03 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_+_164528866 | 0.84 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr3_+_141103634 | 0.79 |
ENST00000507722.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr7_-_150754935 | 0.68 |
ENST00000297518.4
|
CDK5
|
cyclin-dependent kinase 5 |
| chr2_-_208031943 | 0.60 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr2_+_201997595 | 0.49 |
ENST00000470178.2
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr2_+_201997492 | 0.47 |
ENST00000494258.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr2_+_201997676 | 0.47 |
ENST00000462763.1
ENST00000479953.2 |
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr3_+_112051994 | 0.41 |
ENST00000473539.1
ENST00000315711.8 ENST00000383681.3 |
CD200
|
CD200 molecule |
| chr6_+_80341000 | 0.40 |
ENST00000369838.4
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2 |
| chr1_-_214638146 | 0.38 |
ENST00000543945.1
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chrX_+_149531524 | 0.37 |
ENST00000370401.2
|
MAMLD1
|
mastermind-like domain containing 1 |
| chr19_+_12862486 | 0.37 |
ENST00000549706.1
|
BEST2
|
bestrophin 2 |
| chr19_+_12862604 | 0.36 |
ENST00000553030.1
|
BEST2
|
bestrophin 2 |
| chr10_+_99344104 | 0.36 |
ENST00000555577.1
ENST00000370649.3 |
PI4K2A
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha Phosphatidylinositol 4-kinase type 2-alpha; Uncharacterized protein |
| chr6_-_39282329 | 0.35 |
ENST00000373231.4
|
KCNK17
|
potassium channel, subfamily K, member 17 |
| chr6_+_35996859 | 0.35 |
ENST00000472333.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr14_-_53331239 | 0.33 |
ENST00000553663.1
|
FERMT2
|
fermitin family member 2 |
| chr6_-_39282221 | 0.33 |
ENST00000453413.2
|
KCNK17
|
potassium channel, subfamily K, member 17 |
| chr11_+_124932986 | 0.32 |
ENST00000407458.1
ENST00000298280.5 |
SLC37A2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr11_+_124933191 | 0.32 |
ENST00000532000.1
ENST00000308074.4 |
SLC37A2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr11_+_369804 | 0.32 |
ENST00000329962.6
|
B4GALNT4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4 |
| chr16_-_10652993 | 0.30 |
ENST00000536829.1
|
EMP2
|
epithelial membrane protein 2 |
| chr11_+_124932955 | 0.30 |
ENST00000403796.2
|
SLC37A2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr11_-_72432950 | 0.29 |
ENST00000426523.1
ENST00000429686.1 |
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr20_-_56285595 | 0.25 |
ENST00000395816.3
ENST00000347215.4 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr20_-_56286479 | 0.24 |
ENST00000265626.4
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr2_-_25475120 | 0.23 |
ENST00000380746.4
ENST00000402667.1 |
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha |
| chr6_-_133119668 | 0.22 |
ENST00000275227.4
ENST00000538764.1 |
SLC18B1
|
solute carrier family 18, subfamily B, member 1 |
| chr14_+_96722539 | 0.22 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr1_+_207262627 | 0.20 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein, beta |
| chr1_+_207262578 | 0.20 |
ENST00000243611.5
ENST00000367076.3 |
C4BPB
|
complement component 4 binding protein, beta |
| chr21_+_34602200 | 0.20 |
ENST00000382264.3
ENST00000382241.3 ENST00000404220.3 ENST00000342136.4 |
IFNAR2
|
interferon (alpha, beta and omega) receptor 2 |
| chr12_+_98909260 | 0.19 |
ENST00000556029.1
|
TMPO
|
thymopoietin |
| chr8_+_79428539 | 0.19 |
ENST00000352966.5
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr17_-_36358166 | 0.19 |
ENST00000537432.1
|
TBC1D3
|
TBC1 domain family, member 3 |
| chr22_+_25003568 | 0.19 |
ENST00000447416.1
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr1_+_207262540 | 0.18 |
ENST00000452902.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr19_+_507299 | 0.18 |
ENST00000359315.5
|
TPGS1
|
tubulin polyglutamylase complex subunit 1 |
| chr7_-_141957847 | 0.17 |
ENST00000552471.1
ENST00000547058.2 |
PRSS58
|
protease, serine, 58 |
| chr3_-_15382875 | 0.17 |
ENST00000408919.3
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr3_-_9811595 | 0.17 |
ENST00000256460.3
|
CAMK1
|
calcium/calmodulin-dependent protein kinase I |
| chr1_+_207262170 | 0.16 |
ENST00000367078.3
|
C4BPB
|
complement component 4 binding protein, beta |
| chr1_-_52870104 | 0.16 |
ENST00000371568.3
|
ORC1
|
origin recognition complex, subunit 1 |
| chr21_+_34602377 | 0.16 |
ENST00000342101.3
ENST00000413881.1 ENST00000443073.1 |
IFNAR2
|
interferon (alpha, beta and omega) receptor 2 |
| chr1_-_52870059 | 0.15 |
ENST00000371566.1
|
ORC1
|
origin recognition complex, subunit 1 |
| chr5_+_36608422 | 0.15 |
ENST00000381918.3
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr2_+_90153696 | 0.14 |
ENST00000417279.2
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 (gene/pseudogene) |
| chr12_-_10955226 | 0.13 |
ENST00000240687.2
|
TAS2R7
|
taste receptor, type 2, member 7 |
| chr22_+_24999114 | 0.13 |
ENST00000412658.1
ENST00000445029.1 ENST00000419133.1 ENST00000400382.1 ENST00000438643.2 ENST00000452551.1 ENST00000400383.1 ENST00000412898.1 ENST00000400380.1 ENST00000455483.1 ENST00000430289.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr22_+_25003606 | 0.13 |
ENST00000432867.1
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr17_+_66624280 | 0.12 |
ENST00000585484.1
|
RP11-118B18.1
|
RP11-118B18.1 |
| chr6_+_6588902 | 0.12 |
ENST00000230568.4
|
LY86
|
lymphocyte antigen 86 |
| chr12_-_110271178 | 0.12 |
ENST00000261740.2
ENST00000392719.2 ENST00000346520.2 |
TRPV4
|
transient receptor potential cation channel, subfamily V, member 4 |
| chr11_-_2323089 | 0.12 |
ENST00000456145.2
|
C11orf21
|
chromosome 11 open reading frame 21 |
| chr11_+_112130988 | 0.11 |
ENST00000595053.1
|
AP002884.2
|
LOC100132686 protein; Uncharacterized protein |
| chr6_+_44191507 | 0.11 |
ENST00000371724.1
ENST00000371713.1 |
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr1_+_45478568 | 0.11 |
ENST00000428106.1
|
UROD
|
uroporphyrinogen decarboxylase |
| chr7_-_142207004 | 0.11 |
ENST00000426318.2
|
TRBV10-2
|
T cell receptor beta variable 10-2 |
| chrX_-_34675391 | 0.11 |
ENST00000275954.3
|
TMEM47
|
transmembrane protein 47 |
| chr2_+_3642545 | 0.11 |
ENST00000382062.2
ENST00000236693.7 ENST00000349077.4 |
COLEC11
|
collectin sub-family member 11 |
| chr16_+_70328680 | 0.11 |
ENST00000563206.1
ENST00000451014.3 ENST00000568625.1 |
DDX19B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 19B |
| chr11_-_63330842 | 0.11 |
ENST00000255695.1
|
HRASLS2
|
HRAS-like suppressor 2 |
| chr10_+_82297658 | 0.11 |
ENST00000339284.2
|
SH2D4B
|
SH2 domain containing 4B |
| chr12_-_7261772 | 0.10 |
ENST00000545280.1
ENST00000543933.1 ENST00000545337.1 ENST00000544702.1 ENST00000266542.4 |
C1RL
|
complement component 1, r subcomponent-like |
| chr17_+_43922241 | 0.10 |
ENST00000329196.5
|
SPPL2C
|
signal peptide peptidase like 2C |
| chr11_+_67171391 | 0.10 |
ENST00000312390.5
|
TBC1D10C
|
TBC1 domain family, member 10C |
| chr11_+_67171548 | 0.10 |
ENST00000542590.1
|
TBC1D10C
|
TBC1 domain family, member 10C |
| chr19_-_36304201 | 0.10 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr9_-_115095123 | 0.10 |
ENST00000458258.1
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr11_+_67171358 | 0.10 |
ENST00000526387.1
|
TBC1D10C
|
TBC1 domain family, member 10C |
| chr1_-_54405773 | 0.10 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr17_+_45908974 | 0.09 |
ENST00000269025.4
|
LRRC46
|
leucine rich repeat containing 46 |
| chr3_+_123813509 | 0.09 |
ENST00000460856.1
ENST00000240874.3 |
KALRN
|
kalirin, RhoGEF kinase |
| chr19_+_47421933 | 0.08 |
ENST00000404338.3
|
ARHGAP35
|
Rho GTPase activating protein 35 |
| chr8_+_1993152 | 0.08 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr12_+_49717019 | 0.08 |
ENST00000549275.1
ENST00000551245.1 ENST00000380327.5 ENST00000548311.1 ENST00000550346.1 ENST00000550709.1 ENST00000549534.1 ENST00000257909.3 |
TROAP
|
trophinin associated protein |
| chr14_+_96722152 | 0.08 |
ENST00000216629.6
|
BDKRB1
|
bradykinin receptor B1 |
| chr7_-_20256965 | 0.08 |
ENST00000400331.5
ENST00000332878.4 |
MACC1
|
metastasis associated in colon cancer 1 |
| chr19_-_55866061 | 0.08 |
ENST00000588572.2
ENST00000593184.1 ENST00000589467.1 |
COX6B2
|
cytochrome c oxidase subunit VIb polypeptide 2 (testis) |
| chr22_+_25003626 | 0.08 |
ENST00000451366.1
ENST00000406383.2 ENST00000428855.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr17_-_56358287 | 0.08 |
ENST00000225275.3
ENST00000340482.3 |
MPO
|
myeloperoxidase |
| chr16_-_70729496 | 0.08 |
ENST00000567648.1
|
VAC14
|
Vac14 homolog (S. cerevisiae) |
| chr7_-_140714739 | 0.07 |
ENST00000467334.1
ENST00000324787.5 |
MRPS33
|
mitochondrial ribosomal protein S33 |
| chr20_+_58296265 | 0.07 |
ENST00000395636.2
ENST00000361300.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr8_+_1993173 | 0.07 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr17_+_6347729 | 0.07 |
ENST00000572447.1
|
FAM64A
|
family with sequence similarity 64, member A |
| chr20_-_1165117 | 0.07 |
ENST00000381894.3
|
TMEM74B
|
transmembrane protein 74B |
| chr3_-_157251383 | 0.07 |
ENST00000487753.1
ENST00000489602.1 ENST00000461299.1 ENST00000479987.1 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chrX_+_15767971 | 0.07 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr6_-_87804815 | 0.07 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr12_-_25801478 | 0.07 |
ENST00000540106.1
ENST00000445693.1 ENST00000545543.1 ENST00000542224.1 |
IFLTD1
|
intermediate filament tail domain containing 1 |
| chr7_+_76139833 | 0.07 |
ENST00000257632.5
|
UPK3B
|
uroplakin 3B |
| chrX_-_133930285 | 0.06 |
ENST00000486347.1
ENST00000343004.5 |
FAM122B
|
family with sequence similarity 122B |
| chr10_+_103113802 | 0.06 |
ENST00000370187.3
|
BTRC
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr17_+_6347761 | 0.06 |
ENST00000250056.8
ENST00000571373.1 ENST00000570337.2 ENST00000572595.2 ENST00000576056.1 |
FAM64A
|
family with sequence similarity 64, member A |
| chr12_-_57630873 | 0.06 |
ENST00000556732.1
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 |
| chr20_+_3713314 | 0.06 |
ENST00000254963.2
ENST00000542646.1 ENST00000399701.1 |
HSPA12B
|
heat shock 70kD protein 12B |
| chr12_+_53848549 | 0.06 |
ENST00000439930.3
ENST00000548933.1 ENST00000562264.1 |
PCBP2
|
poly(rC) binding protein 2 |
| chr10_+_103113840 | 0.05 |
ENST00000393441.4
ENST00000408038.2 |
BTRC
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr4_-_40632605 | 0.05 |
ENST00000514014.1
|
RBM47
|
RNA binding motif protein 47 |
| chr10_+_118350468 | 0.05 |
ENST00000358834.4
ENST00000528052.1 ENST00000442761.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr7_+_140378955 | 0.05 |
ENST00000473512.1
|
ADCK2
|
aarF domain containing kinase 2 |
| chr7_+_110731062 | 0.05 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr7_-_140714430 | 0.05 |
ENST00000393008.3
|
MRPS33
|
mitochondrial ribosomal protein S33 |
| chr9_+_125376948 | 0.05 |
ENST00000297913.2
|
OR1Q1
|
olfactory receptor, family 1, subfamily Q, member 1 |
| chrX_+_70364667 | 0.04 |
ENST00000536169.1
ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3
|
neuroligin 3 |
| chr2_+_210444142 | 0.04 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chr10_+_118350522 | 0.04 |
ENST00000530319.1
ENST00000527980.1 ENST00000471549.1 ENST00000534537.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr6_+_134210243 | 0.04 |
ENST00000367882.4
|
TCF21
|
transcription factor 21 |
| chr17_+_11924129 | 0.03 |
ENST00000353533.5
ENST00000415385.3 |
MAP2K4
|
mitogen-activated protein kinase kinase 4 |
| chr2_+_204732487 | 0.03 |
ENST00000302823.3
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr20_+_123010 | 0.03 |
ENST00000382398.3
|
DEFB126
|
defensin, beta 126 |
| chr2_+_231860830 | 0.03 |
ENST00000424440.1
ENST00000452881.1 ENST00000433428.2 ENST00000455816.1 ENST00000440792.1 ENST00000423134.1 |
SPATA3
|
spermatogenesis associated 3 |
| chr14_-_65409438 | 0.03 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr19_+_36359341 | 0.03 |
ENST00000221891.4
|
APLP1
|
amyloid beta (A4) precursor-like protein 1 |
| chr2_+_98262497 | 0.03 |
ENST00000258424.2
|
COX5B
|
cytochrome c oxidase subunit Vb |
| chr14_+_24422795 | 0.03 |
ENST00000313250.5
ENST00000558263.1 ENST00000543741.2 ENST00000421831.1 ENST00000397073.2 ENST00000308178.8 ENST00000382761.3 ENST00000397075.3 ENST00000397074.3 ENST00000559632.1 ENST00000558581.1 |
DHRS4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr1_-_32210275 | 0.03 |
ENST00000440175.2
|
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr17_-_64216748 | 0.02 |
ENST00000585162.1
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr2_-_231860596 | 0.02 |
ENST00000441063.1
ENST00000434094.1 ENST00000418330.1 ENST00000457803.1 ENST00000414876.1 ENST00000446741.1 ENST00000426904.1 |
AC105344.2
|
SPATA3 antisense RNA 1 (head to head) |
| chr4_-_25161996 | 0.02 |
ENST00000513285.1
ENST00000382103.2 |
SEPSECS
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr17_-_43025005 | 0.02 |
ENST00000587309.1
ENST00000593135.1 ENST00000339151.4 |
KIF18B
|
kinesin family member 18B |
| chr17_-_61781750 | 0.02 |
ENST00000582026.1
|
STRADA
|
STE20-related kinase adaptor alpha |
| chr12_+_68042517 | 0.02 |
ENST00000393555.3
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr12_+_53848505 | 0.01 |
ENST00000552819.1
ENST00000455667.3 |
PCBP2
|
poly(rC) binding protein 2 |
| chr19_-_13044494 | 0.01 |
ENST00000593021.1
ENST00000587981.1 ENST00000423140.2 ENST00000314606.4 |
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chr1_-_52344471 | 0.01 |
ENST00000352171.7
ENST00000354831.7 |
NRD1
|
nardilysin (N-arginine dibasic convertase) |
| chr12_-_68647281 | 0.01 |
ENST00000328087.4
ENST00000538666.1 |
IL22
|
interleukin 22 |
| chr14_-_74892805 | 0.01 |
ENST00000331628.3
ENST00000554953.1 |
SYNDIG1L
|
synapse differentiation inducing 1-like |
| chr2_-_42588338 | 0.01 |
ENST00000234301.2
|
COX7A2L
|
cytochrome c oxidase subunit VIIa polypeptide 2 like |
| chr14_-_65409502 | 0.01 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr11_-_2323290 | 0.00 |
ENST00000381153.3
|
C11orf21
|
chromosome 11 open reading frame 21 |
| chr12_+_67663056 | 0.00 |
ENST00000545606.1
|
CAND1
|
cullin-associated and neddylation-dissociated 1 |
| chr14_+_74083548 | 0.00 |
ENST00000381139.1
|
ACOT6
|
acyl-CoA thioesterase 6 |
| chr10_-_104211294 | 0.00 |
ENST00000239125.1
|
C10orf95
|
chromosome 10 open reading frame 95 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HAND1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 1.4 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.4 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.1 | 0.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.9 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.7 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.1 | 0.3 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.3 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.1 | 0.5 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:2000340 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) positive regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000340) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.3 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.8 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.4 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.2 | GO:0018095 | sperm axoneme assembly(GO:0007288) protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.4 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.4 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.0 | GO:0060435 | branchiomeric skeletal muscle development(GO:0014707) bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.3 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.0 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.0 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 1.4 | GO:0097342 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.1 | 0.3 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.9 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.4 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.4 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.1 | 0.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 1.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |