Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
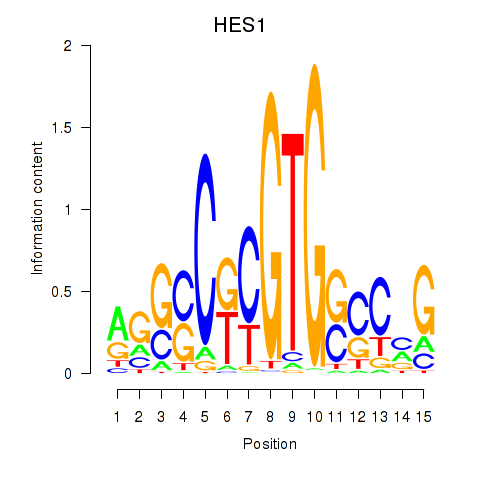
Results for HES1
Z-value: 0.93
Transcription factors associated with HES1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HES1
|
ENSG00000114315.3 | hes family bHLH transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HES1 | hg19_v2_chr3_+_193853927_193853944 | -0.68 | 2.0e-04 | Click! |
Activity profile of HES1 motif
Sorted Z-values of HES1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_89182178 | 3.65 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr15_+_89182156 | 3.60 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr15_+_89181974 | 3.16 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr2_-_225907150 | 2.63 |
ENST00000258390.7
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr3_-_127541679 | 2.48 |
ENST00000265052.5
|
MGLL
|
monoglyceride lipase |
| chr11_-_60719213 | 2.18 |
ENST00000227880.3
|
SLC15A3
|
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr1_-_9189229 | 2.01 |
ENST00000377411.4
|
GPR157
|
G protein-coupled receptor 157 |
| chr7_+_73703728 | 2.01 |
ENST00000361545.5
ENST00000223398.6 |
CLIP2
|
CAP-GLY domain containing linker protein 2 |
| chr16_+_2039946 | 1.85 |
ENST00000248121.2
ENST00000568896.1 |
SYNGR3
|
synaptogyrin 3 |
| chr1_+_23037323 | 1.80 |
ENST00000544305.1
ENST00000374630.3 ENST00000400191.3 ENST00000374632.3 |
EPHB2
|
EPH receptor B2 |
| chr7_-_47621736 | 1.74 |
ENST00000311160.9
|
TNS3
|
tensin 3 |
| chr2_+_150187020 | 1.70 |
ENST00000334166.4
|
LYPD6
|
LY6/PLAUR domain containing 6 |
| chr22_-_50963976 | 1.68 |
ENST00000252785.3
ENST00000395693.3 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr3_-_127542021 | 1.67 |
ENST00000434178.2
|
MGLL
|
monoglyceride lipase |
| chr3_-_127542051 | 1.65 |
ENST00000398104.1
|
MGLL
|
monoglyceride lipase |
| chr22_-_50964558 | 1.40 |
ENST00000535425.1
ENST00000439934.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr6_-_143266297 | 1.39 |
ENST00000367603.2
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr16_-_72127456 | 1.35 |
ENST00000562153.1
|
TXNL4B
|
thioredoxin-like 4B |
| chr1_-_205290865 | 1.35 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chr21_+_34697209 | 1.29 |
ENST00000270139.3
|
IFNAR1
|
interferon (alpha, beta and omega) receptor 1 |
| chr10_-_105615164 | 1.21 |
ENST00000355946.2
ENST00000369774.4 |
SH3PXD2A
|
SH3 and PX domains 2A |
| chr7_-_558876 | 1.20 |
ENST00000354513.5
ENST00000402802.3 |
PDGFA
|
platelet-derived growth factor alpha polypeptide |
| chr1_+_165797024 | 1.18 |
ENST00000372212.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr8_-_142011036 | 1.16 |
ENST00000520892.1
|
PTK2
|
protein tyrosine kinase 2 |
| chr2_+_102759199 | 1.11 |
ENST00000409288.1
ENST00000410023.1 |
IL1R1
|
interleukin 1 receptor, type I |
| chr6_-_154831779 | 1.08 |
ENST00000607772.1
|
CNKSR3
|
CNKSR family member 3 |
| chr2_-_25142708 | 1.06 |
ENST00000260600.5
ENST00000435135.1 |
ADCY3
|
adenylate cyclase 3 |
| chr21_+_45285050 | 1.05 |
ENST00000291572.8
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr7_+_151038785 | 1.04 |
ENST00000413040.2
ENST00000568733.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr22_-_50964849 | 1.02 |
ENST00000543927.1
ENST00000423348.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr17_-_1394940 | 1.01 |
ENST00000570984.2
ENST00000361007.2 |
MYO1C
|
myosin IC |
| chr8_+_145064233 | 1.01 |
ENST00000529301.1
ENST00000395068.4 |
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr22_-_31503490 | 1.00 |
ENST00000400299.2
|
SELM
|
Selenoprotein M |
| chr20_+_6748311 | 0.98 |
ENST00000378827.4
|
BMP2
|
bone morphogenetic protein 2 |
| chr8_+_145064215 | 0.96 |
ENST00000313269.5
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr19_+_42724423 | 0.96 |
ENST00000301215.3
ENST00000597945.1 |
ZNF526
|
zinc finger protein 526 |
| chr11_-_67981295 | 0.96 |
ENST00000405515.1
|
SUV420H1
|
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chr22_+_29279552 | 0.96 |
ENST00000544604.2
|
ZNRF3
|
zinc and ring finger 3 |
| chr2_-_10588630 | 0.95 |
ENST00000234111.4
|
ODC1
|
ornithine decarboxylase 1 |
| chr6_+_44095347 | 0.94 |
ENST00000323267.6
|
TMEM63B
|
transmembrane protein 63B |
| chr3_+_133292851 | 0.93 |
ENST00000503932.1
|
CDV3
|
CDV3 homolog (mouse) |
| chr8_+_104384616 | 0.93 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr16_-_54963026 | 0.91 |
ENST00000560208.1
ENST00000557792.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr10_+_103892787 | 0.91 |
ENST00000278070.2
ENST00000413464.2 |
PPRC1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr6_-_43543702 | 0.90 |
ENST00000265351.7
|
XPO5
|
exportin 5 |
| chr1_-_111743285 | 0.89 |
ENST00000357640.4
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr10_+_90750493 | 0.89 |
ENST00000357339.2
ENST00000355279.2 |
FAS
|
Fas cell surface death receptor |
| chr4_+_1873155 | 0.88 |
ENST00000507820.1
ENST00000514045.1 |
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1 |
| chr17_+_74381343 | 0.86 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr3_+_133292574 | 0.86 |
ENST00000264993.3
|
CDV3
|
CDV3 homolog (mouse) |
| chr7_-_131241361 | 0.85 |
ENST00000378555.3
ENST00000322985.9 ENST00000541194.1 ENST00000537928.1 |
PODXL
|
podocalyxin-like |
| chr6_+_37787704 | 0.85 |
ENST00000474522.1
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr10_+_90750378 | 0.85 |
ENST00000355740.2
ENST00000352159.4 |
FAS
|
Fas cell surface death receptor |
| chr8_-_141645645 | 0.83 |
ENST00000519980.1
ENST00000220592.5 |
AGO2
|
argonaute RISC catalytic component 2 |
| chr19_+_14544099 | 0.82 |
ENST00000242783.6
ENST00000586557.1 ENST00000590097.1 |
PKN1
|
protein kinase N1 |
| chr14_+_93799556 | 0.81 |
ENST00000256339.4
|
UNC79
|
unc-79 homolog (C. elegans) |
| chr21_-_44495919 | 0.79 |
ENST00000398158.1
|
CBS
|
cystathionine-beta-synthase |
| chr2_-_136743169 | 0.78 |
ENST00000264161.4
|
DARS
|
aspartyl-tRNA synthetase |
| chr1_+_41445413 | 0.78 |
ENST00000541520.1
|
CTPS1
|
CTP synthase 1 |
| chr17_-_48207157 | 0.78 |
ENST00000330175.4
ENST00000503131.1 |
SAMD14
|
sterile alpha motif domain containing 14 |
| chr17_-_28257080 | 0.77 |
ENST00000579954.1
ENST00000540801.1 ENST00000269033.3 ENST00000590153.1 ENST00000582084.1 |
SSH2
|
slingshot protein phosphatase 2 |
| chr5_+_156693091 | 0.77 |
ENST00000318218.6
ENST00000442283.2 ENST00000522463.1 ENST00000521420.1 |
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr2_+_219433588 | 0.76 |
ENST00000295701.5
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr17_+_46985731 | 0.76 |
ENST00000360943.5
|
UBE2Z
|
ubiquitin-conjugating enzyme E2Z |
| chr2_-_136743039 | 0.76 |
ENST00000537273.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr19_+_33072373 | 0.76 |
ENST00000586035.1
|
PDCD5
|
programmed cell death 5 |
| chr11_+_65029421 | 0.75 |
ENST00000541089.1
|
POLA2
|
polymerase (DNA directed), alpha 2, accessory subunit |
| chr20_+_61273797 | 0.74 |
ENST00000217159.1
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1 |
| chr8_+_32406179 | 0.74 |
ENST00000405005.3
|
NRG1
|
neuregulin 1 |
| chr20_+_55966444 | 0.72 |
ENST00000356208.5
ENST00000440234.2 |
RBM38
|
RNA binding motif protein 38 |
| chr15_-_58357932 | 0.70 |
ENST00000347587.3
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr3_-_47823298 | 0.69 |
ENST00000254480.5
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chr17_+_7348658 | 0.69 |
ENST00000570557.1
ENST00000536404.2 ENST00000576360.1 |
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr11_-_9286921 | 0.68 |
ENST00000328194.3
|
DENND5A
|
DENN/MADD domain containing 5A |
| chr17_-_79849438 | 0.68 |
ENST00000331204.4
ENST00000505490.2 |
ALYREF
|
Aly/REF export factor |
| chr5_+_156693159 | 0.68 |
ENST00000347377.6
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr22_+_38302285 | 0.68 |
ENST00000215957.6
|
MICALL1
|
MICAL-like 1 |
| chr16_-_72127550 | 0.67 |
ENST00000268483.3
|
TXNL4B
|
thioredoxin-like 4B |
| chrX_-_9734004 | 0.67 |
ENST00000467482.1
ENST00000380929.2 |
GPR143
|
G protein-coupled receptor 143 |
| chr17_+_7348374 | 0.67 |
ENST00000306071.2
ENST00000572857.1 |
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr15_-_58357866 | 0.67 |
ENST00000537372.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr9_+_33025209 | 0.65 |
ENST00000330899.4
ENST00000544625.1 |
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1 |
| chr12_-_89919965 | 0.65 |
ENST00000548729.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr1_+_44445549 | 0.65 |
ENST00000356836.6
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr19_+_49622646 | 0.65 |
ENST00000334186.4
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr19_+_984313 | 0.65 |
ENST00000251289.5
ENST00000587001.2 ENST00000607440.1 |
WDR18
|
WD repeat domain 18 |
| chr10_-_62493223 | 0.64 |
ENST00000373827.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr16_-_54962625 | 0.64 |
ENST00000559432.1
|
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr19_+_33071974 | 0.63 |
ENST00000590247.2
ENST00000419343.3 ENST00000592786.1 ENST00000379316.3 |
PDCD5
|
programmed cell death 5 |
| chr1_-_167522982 | 0.63 |
ENST00000370509.4
|
CREG1
|
cellular repressor of E1A-stimulated genes 1 |
| chr6_+_31588478 | 0.63 |
ENST00000376007.4
ENST00000376033.2 |
PRRC2A
|
proline-rich coiled-coil 2A |
| chr1_+_44444865 | 0.62 |
ENST00000372324.1
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr16_-_4466622 | 0.62 |
ENST00000570645.1
ENST00000574025.1 ENST00000572898.1 ENST00000537233.2 ENST00000571059.1 ENST00000251166.4 |
CORO7
|
coronin 7 |
| chr1_-_6662919 | 0.62 |
ENST00000377658.4
ENST00000377663.3 |
KLHL21
|
kelch-like family member 21 |
| chr8_+_141521386 | 0.61 |
ENST00000220913.5
ENST00000519533.1 |
CHRAC1
|
chromatin accessibility complex 1 |
| chr8_-_145515055 | 0.60 |
ENST00000526552.1
ENST00000529231.1 ENST00000307404.5 |
BOP1
|
block of proliferation 1 |
| chr7_+_100303676 | 0.59 |
ENST00000303151.4
|
POP7
|
processing of precursor 7, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr12_+_7023491 | 0.59 |
ENST00000541477.1
ENST00000229277.1 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr10_+_81107271 | 0.59 |
ENST00000448165.1
|
PPIF
|
peptidylprolyl isomerase F |
| chr1_-_231004220 | 0.59 |
ENST00000366663.5
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr6_+_44094627 | 0.59 |
ENST00000259746.9
|
TMEM63B
|
transmembrane protein 63B |
| chr7_-_139477500 | 0.58 |
ENST00000406875.3
ENST00000428878.2 |
HIPK2
|
homeodomain interacting protein kinase 2 |
| chr17_+_7255208 | 0.58 |
ENST00000333751.3
|
KCTD11
|
potassium channel tetramerization domain containing 11 |
| chr3_+_9773409 | 0.58 |
ENST00000433861.2
ENST00000424362.1 ENST00000383829.2 ENST00000302054.3 ENST00000420291.1 |
BRPF1
|
bromodomain and PHD finger containing, 1 |
| chr16_-_54962704 | 0.57 |
ENST00000502066.2
ENST00000560912.1 ENST00000558952.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr3_-_179754806 | 0.57 |
ENST00000485199.1
|
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr13_-_102068706 | 0.57 |
ENST00000251127.6
|
NALCN
|
sodium leak channel, non-selective |
| chr8_+_31496809 | 0.56 |
ENST00000518104.1
ENST00000519301.1 |
NRG1
|
neuregulin 1 |
| chr3_+_133292759 | 0.56 |
ENST00000431519.2
|
CDV3
|
CDV3 homolog (mouse) |
| chr21_-_40685536 | 0.55 |
ENST00000341322.4
|
BRWD1
|
bromodomain and WD repeat domain containing 1 |
| chr2_+_178077477 | 0.55 |
ENST00000411529.2
ENST00000435711.1 |
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr17_+_45726803 | 0.55 |
ENST00000535458.2
ENST00000583648.1 |
KPNB1
|
karyopherin (importin) beta 1 |
| chr15_+_52311398 | 0.55 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr9_-_140484917 | 0.55 |
ENST00000298585.2
|
ZMYND19
|
zinc finger, MYND-type containing 19 |
| chr11_+_94277017 | 0.54 |
ENST00000358752.2
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chr1_-_38273840 | 0.54 |
ENST00000373044.2
|
YRDC
|
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr12_+_107349497 | 0.54 |
ENST00000548125.1
ENST00000280756.4 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chr10_-_90751038 | 0.54 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr19_-_1863567 | 0.53 |
ENST00000250916.4
|
KLF16
|
Kruppel-like factor 16 |
| chr11_-_46142615 | 0.53 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr21_-_18985230 | 0.53 |
ENST00000457956.1
ENST00000348354.6 |
BTG3
|
BTG family, member 3 |
| chr10_-_121302195 | 0.53 |
ENST00000369103.2
|
RGS10
|
regulator of G-protein signaling 10 |
| chr17_-_74722536 | 0.53 |
ENST00000585429.1
|
JMJD6
|
jumonji domain containing 6 |
| chr2_+_201390843 | 0.53 |
ENST00000357799.4
ENST00000409203.3 |
SGOL2
|
shugoshin-like 2 (S. pombe) |
| chr16_-_54962415 | 0.53 |
ENST00000501177.3
ENST00000559598.2 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr20_-_43280325 | 0.52 |
ENST00000537820.1
|
ADA
|
adenosine deaminase |
| chr19_+_489140 | 0.52 |
ENST00000587541.1
|
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr21_+_45138941 | 0.52 |
ENST00000398081.1
ENST00000468090.1 ENST00000291565.4 |
PDXK
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr12_-_109251345 | 0.51 |
ENST00000360239.3
ENST00000326495.5 ENST00000551165.1 |
SSH1
|
slingshot protein phosphatase 1 |
| chr6_+_43543864 | 0.51 |
ENST00000372236.4
ENST00000535400.1 |
POLH
|
polymerase (DNA directed), eta |
| chr1_-_23670817 | 0.51 |
ENST00000478691.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr17_-_78450398 | 0.51 |
ENST00000306773.4
|
NPTX1
|
neuronal pentraxin I |
| chr2_+_106361333 | 0.50 |
ENST00000233154.4
ENST00000451463.2 |
NCK2
|
NCK adaptor protein 2 |
| chr12_-_106641728 | 0.50 |
ENST00000378026.4
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr11_-_65430251 | 0.50 |
ENST00000534283.1
ENST00000527749.1 ENST00000533187.1 ENST00000525693.1 ENST00000534558.1 ENST00000532879.1 ENST00000532999.1 |
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr2_+_192543153 | 0.49 |
ENST00000425611.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr17_+_79650962 | 0.49 |
ENST00000329138.4
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr14_+_94577074 | 0.49 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr12_+_77158021 | 0.49 |
ENST00000550876.1
|
ZDHHC17
|
zinc finger, DHHC-type containing 17 |
| chr2_+_192542850 | 0.49 |
ENST00000410026.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr17_+_36861735 | 0.48 |
ENST00000378137.5
ENST00000325718.7 |
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr19_+_10982336 | 0.48 |
ENST00000344150.4
|
CARM1
|
coactivator-associated arginine methyltransferase 1 |
| chr7_-_80548667 | 0.48 |
ENST00000265361.3
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr17_+_28256874 | 0.48 |
ENST00000541045.1
ENST00000536908.2 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr6_+_31126291 | 0.48 |
ENST00000376257.3
ENST00000376255.4 |
TCF19
|
transcription factor 19 |
| chr12_+_7023735 | 0.48 |
ENST00000538763.1
ENST00000544774.1 ENST00000545045.2 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr11_-_615570 | 0.47 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr10_-_99094458 | 0.47 |
ENST00000371019.2
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2 |
| chr1_-_43424500 | 0.47 |
ENST00000415851.2
ENST00000426263.3 ENST00000372500.3 |
SLC2A1
|
solute carrier family 2 (facilitated glucose transporter), member 1 |
| chr14_-_53162361 | 0.47 |
ENST00000395686.3
|
ERO1L
|
ERO1-like (S. cerevisiae) |
| chr7_-_92463210 | 0.47 |
ENST00000265734.4
|
CDK6
|
cyclin-dependent kinase 6 |
| chr6_-_13814663 | 0.47 |
ENST00000359495.2
ENST00000379170.4 |
MCUR1
|
mitochondrial calcium uniporter regulator 1 |
| chr16_-_4466565 | 0.46 |
ENST00000572467.1
ENST00000423908.2 ENST00000572044.1 ENST00000571052.1 |
CORO7-PAM16
CORO7
|
CORO7-PAM16 readthrough coronin 7 |
| chr3_+_127348005 | 0.46 |
ENST00000342480.6
|
PODXL2
|
podocalyxin-like 2 |
| chr14_-_77279153 | 0.46 |
ENST00000251089.2
|
ANGEL1
|
angel homolog 1 (Drosophila) |
| chr2_-_224702257 | 0.46 |
ENST00000409375.1
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit |
| chr12_-_89920030 | 0.45 |
ENST00000413530.1
ENST00000547474.1 |
GALNT4
POC1B-GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr14_+_103243813 | 0.45 |
ENST00000560371.1
ENST00000347662.4 ENST00000392745.2 ENST00000539721.1 ENST00000560463.1 |
TRAF3
|
TNF receptor-associated factor 3 |
| chr7_-_130353553 | 0.45 |
ENST00000330992.7
ENST00000445977.2 |
COPG2
|
coatomer protein complex, subunit gamma 2 |
| chr13_+_76123883 | 0.45 |
ENST00000377595.3
|
UCHL3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr2_-_120124258 | 0.44 |
ENST00000409877.1
ENST00000409523.1 ENST00000409466.2 ENST00000414534.1 |
C2orf76
|
chromosome 2 open reading frame 76 |
| chr12_+_26111823 | 0.44 |
ENST00000381352.3
ENST00000535907.1 ENST00000405154.2 |
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr9_+_92219919 | 0.44 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr2_-_135476552 | 0.44 |
ENST00000281924.6
|
TMEM163
|
transmembrane protein 163 |
| chr1_+_41707996 | 0.44 |
ENST00000425554.1
|
RP11-399E6.1
|
RP11-399E6.1 |
| chr19_-_38397285 | 0.44 |
ENST00000303868.5
|
WDR87
|
WD repeat domain 87 |
| chrX_+_153770421 | 0.44 |
ENST00000369609.5
ENST00000369607.1 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr17_-_73257667 | 0.44 |
ENST00000538886.1
ENST00000580799.1 ENST00000351904.7 ENST00000537686.1 |
GGA3
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr3_-_179754556 | 0.43 |
ENST00000263962.8
|
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr8_-_72756667 | 0.43 |
ENST00000325509.4
|
MSC
|
musculin |
| chr19_+_10982189 | 0.43 |
ENST00000327064.4
ENST00000588947.1 |
CARM1
|
coactivator-associated arginine methyltransferase 1 |
| chr21_+_45527171 | 0.43 |
ENST00000291576.7
ENST00000456705.1 |
PWP2
|
PWP2 periodic tryptophan protein homolog (yeast) |
| chr5_+_151151504 | 0.43 |
ENST00000356245.3
ENST00000507878.2 |
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr16_-_4401258 | 0.42 |
ENST00000577031.1
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr16_-_4401284 | 0.42 |
ENST00000318059.3
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr22_-_45636650 | 0.42 |
ENST00000336156.5
|
KIAA0930
|
KIAA0930 |
| chr8_+_38614807 | 0.42 |
ENST00000330691.6
ENST00000348567.4 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr7_-_140340576 | 0.42 |
ENST00000275884.6
ENST00000475837.1 |
DENND2A
|
DENN/MADD domain containing 2A |
| chr5_+_6633534 | 0.42 |
ENST00000537411.1
ENST00000538824.1 |
SRD5A1
|
steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) |
| chr3_+_130613226 | 0.41 |
ENST00000509662.1
ENST00000328560.8 ENST00000428331.2 ENST00000359644.3 ENST00000422190.2 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr5_+_151151471 | 0.41 |
ENST00000394123.3
ENST00000543466.1 |
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr17_-_30185971 | 0.41 |
ENST00000378634.2
|
COPRS
|
coordinator of PRMT5, differentiation stimulator |
| chr1_-_23670752 | 0.41 |
ENST00000302271.6
ENST00000426846.2 ENST00000427764.2 ENST00000606561.1 ENST00000374616.3 |
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr14_-_67982146 | 0.40 |
ENST00000557779.1
ENST00000557006.1 |
TMEM229B
|
transmembrane protein 229B |
| chr21_-_43373999 | 0.40 |
ENST00000380486.3
|
C2CD2
|
C2 calcium-dependent domain containing 2 |
| chr14_-_95236551 | 0.40 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr2_-_120124383 | 0.40 |
ENST00000334816.7
|
C2orf76
|
chromosome 2 open reading frame 76 |
| chr7_-_2354099 | 0.40 |
ENST00000222990.3
|
SNX8
|
sorting nexin 8 |
| chr6_-_43197189 | 0.40 |
ENST00000509253.1
ENST00000393987.2 ENST00000230431.6 |
DNPH1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr1_-_19283163 | 0.39 |
ENST00000455833.2
|
IFFO2
|
intermediate filament family orphan 2 |
| chr2_-_224702201 | 0.39 |
ENST00000446015.2
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit |
| chr16_-_87417351 | 0.39 |
ENST00000311635.7
|
FBXO31
|
F-box protein 31 |
| chr22_-_39151463 | 0.39 |
ENST00000405510.1
ENST00000433561.1 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr3_-_170744498 | 0.39 |
ENST00000382808.4
ENST00000314251.3 |
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr5_-_6633397 | 0.39 |
ENST00000264670.6
ENST00000539938.1 |
NSUN2
|
NOP2/Sun RNA methyltransferase family, member 2 |
| chr1_+_28844778 | 0.38 |
ENST00000411533.1
|
RCC1
|
regulator of chromosome condensation 1 |
| chr10_-_79397391 | 0.38 |
ENST00000286628.8
ENST00000406533.3 ENST00000354353.5 ENST00000404857.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr22_+_45559722 | 0.38 |
ENST00000347635.4
ENST00000407019.2 ENST00000424634.1 ENST00000417702.1 ENST00000425733.2 ENST00000430547.1 |
NUP50
|
nucleoporin 50kDa |
| chr8_+_1922024 | 0.38 |
ENST00000320248.3
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11 |
| chr4_-_926161 | 0.38 |
ENST00000511163.1
|
GAK
|
cyclin G associated kinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of HES1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.8 | 5.8 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.5 | 1.5 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.4 | 1.2 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.4 | 1.1 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.4 | 1.1 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.4 | 1.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.3 | 1.4 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.3 | 1.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.3 | 0.9 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.3 | 1.2 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.3 | 1.7 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.3 | 0.9 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.3 | 0.8 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.3 | 0.8 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.3 | 0.8 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.2 | 1.2 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.2 | 1.0 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.2 | 0.9 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.2 | 0.4 | GO:0061054 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.2 | 0.9 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.2 | 0.9 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.2 | 0.8 | GO:0046102 | inosine metabolic process(GO:0046102) |
| 0.2 | 1.2 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.2 | 0.4 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 0.2 | 1.8 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.2 | 0.6 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.2 | 4.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.2 | 2.6 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.2 | 1.3 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.2 | 0.5 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 0.5 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.2 | 0.5 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.2 | 0.5 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.2 | 1.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 0.7 | GO:0006726 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) regulation of melanosome transport(GO:1902908) |
| 0.2 | 0.7 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.2 | 0.6 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 1.4 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 0.4 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.1 | 1.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.4 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.8 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.8 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.9 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 1.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 0.4 | GO:1905075 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.1 | 0.6 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.1 | 2.0 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.1 | 0.9 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.1 | 0.5 | GO:0014040 | positive regulation of Schwann cell differentiation(GO:0014040) |
| 0.1 | 1.4 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.5 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.4 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.1 | 0.5 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 0.4 | GO:0035602 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) mammary gland specification(GO:0060594) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.1 | 0.6 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 | 0.5 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 0.1 | 0.9 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 0.3 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 0.4 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.1 | 0.3 | GO:1905068 | arterial endothelial cell fate commitment(GO:0060844) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.6 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.1 | 1.1 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 0.6 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.4 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.1 | 0.5 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 | 0.4 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.3 | GO:0044111 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) development involved in symbiotic interaction(GO:0044111) |
| 0.1 | 0.5 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.1 | 0.2 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.1 | 0.6 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.4 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.1 | 0.1 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.1 | 0.7 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 0.3 | GO:0019605 | butyrate metabolic process(GO:0019605) |
| 0.1 | 0.5 | GO:0045007 | depurination(GO:0045007) |
| 0.1 | 0.4 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 2.0 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 0.1 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.1 | 0.5 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 0.9 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.7 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.1 | 0.6 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.1 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.4 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.3 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.1 | 0.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.5 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.1 | 0.2 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.2 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.1 | 0.6 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 0.2 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.1 | 0.2 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 0.1 | 0.2 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.1 | 0.2 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.1 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 0.4 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.1 | 1.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 | 0.3 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 | 0.4 | GO:0072364 | regulation of cellular ketone metabolic process by regulation of transcription from RNA polymerase II promoter(GO:0072364) |
| 0.0 | 0.2 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.2 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.7 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.7 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.4 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.5 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.2 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.0 | 0.5 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.5 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.3 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.4 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.3 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.2 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.8 | GO:0032780 | protein import into mitochondrial matrix(GO:0030150) negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.3 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.4 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.3 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.6 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 1.0 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 1.2 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.7 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.6 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.9 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.2 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.0 | 0.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.2 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.3 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 1.7 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.4 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.4 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.0 | 0.6 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 1.0 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.1 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.8 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.4 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.6 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 1.0 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.0 | 0.3 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.3 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.2 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.3 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 1.5 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 1.1 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.5 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.0 | 0.2 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.1 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.6 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0050975 | sensory perception of touch(GO:0050975) detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.2 | GO:0071499 | cellular response to laminar fluid shear stress(GO:0071499) |
| 0.0 | 0.7 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 2.3 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.2 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 0.0 | 0.1 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 1.8 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.2 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 1.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 1.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.3 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.1 | GO:2000619 | regulation of histone H4-K16 acetylation(GO:2000618) negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.1 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.0 | 0.1 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.6 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.3 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 1.7 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.3 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.8 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 1.0 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.4 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.1 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.3 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.2 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.1 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.1 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.0 | 0.9 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 0.0 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.0 | 0.1 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.0 | 0.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.0 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.5 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.3 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.4 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.0 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.0 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.3 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.3 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.2 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.0 | GO:0038001 | paracrine signaling(GO:0038001) corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.0 | 0.3 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.3 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.4 | GO:1900115 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.5 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.7 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.2 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.0 | 0.0 | GO:0000429 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.0 | 0.0 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 2.1 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 3.8 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.4 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.4 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.1 | GO:0032328 | alanine transport(GO:0032328) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.3 | 1.0 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.2 | 1.0 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 0.8 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.2 | 0.2 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.2 | 0.5 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.2 | 0.5 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.2 | 0.6 | GO:1990742 | microvesicle(GO:1990742) |
| 0.2 | 0.6 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.2 | 1.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 1.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 11.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.6 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.6 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 0.9 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 2.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 0.6 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.6 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 1.7 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 1.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.9 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.2 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 0.7 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 0.2 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.1 | 0.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 2.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.4 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 1.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.4 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.6 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.8 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.4 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.2 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 1.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.7 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.5 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.2 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.7 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.2 | GO:0044326 | chromaffin granule membrane(GO:0042584) dendritic spine neck(GO:0044326) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.5 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 3.0 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 2.0 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.7 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 1.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 1.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 1.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.5 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 3.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.5 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 5.0 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 5.4 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 1.1 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.7 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 1.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.2 | GO:0035578 | azurophil granule lumen(GO:0035578) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.4 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.6 | 1.7 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.5 | 2.2 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.5 | 1.4 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.4 | 1.2 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.4 | 4.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.4 | 1.1 | GO:0050421 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.4 | 1.1 | GO:0019961 | interferon binding(GO:0019961) |
| 0.3 | 1.7 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.3 | 5.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 1.3 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.3 | 1.5 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.3 | 0.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.3 | 0.8 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.3 | 1.1 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.3 | 0.8 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.3 | 0.8 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 0.9 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.2 | 0.5 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.2 | 0.5 | GO:0008478 | pyridoxal kinase activity(GO:0008478) lithium ion binding(GO:0031403) |
| 0.2 | 1.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 0.5 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.2 | 1.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.6 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.7 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 0.9 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.8 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.7 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 1.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.5 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 0.5 | GO:0032406 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.1 | 0.7 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.4 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 0.6 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 1.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 1.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.3 | GO:0043337 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.1 | 0.5 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 1.0 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.6 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.3 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.1 | 0.4 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 1.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 1.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.3 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.9 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 1.6 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 2.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.4 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.6 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.5 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.3 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 0.1 | 0.3 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.3 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 0.2 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.1 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.6 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.7 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.7 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 0.4 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 1.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.2 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.1 | 0.2 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.2 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.1 | 0.5 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 1.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.6 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.5 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.9 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 1.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0050816 | phosphoserine binding(GO:0050815) phosphothreonine binding(GO:0050816) |
| 0.0 | 0.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.5 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 2.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.4 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.4 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.4 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 1.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.7 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 1.0 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.9 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.1 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.7 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.5 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 1.4 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 1.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.7 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 1.0 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 1.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 2.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.5 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 3.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.4 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.0 | 0.1 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.0 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.4 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.2 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 1.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 2.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 2.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 2.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.5 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.7 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 1.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.0 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 12.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 1.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 1.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 1.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.6 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 1.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.7 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 0.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.5 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.7 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 2.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.0 | 1.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 1.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 3.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.1 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.2 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.4 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |