Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for HLF_TEF
Z-value: 0.62
Transcription factors associated with HLF_TEF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
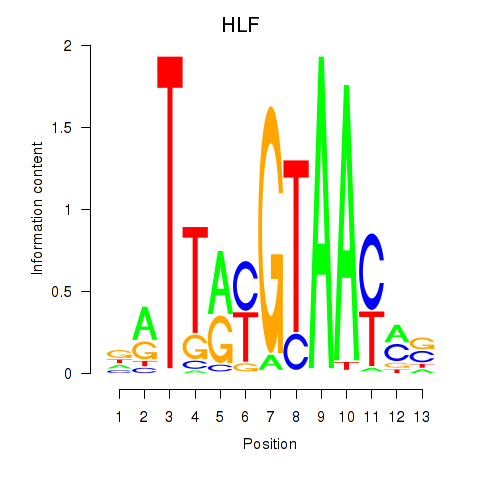
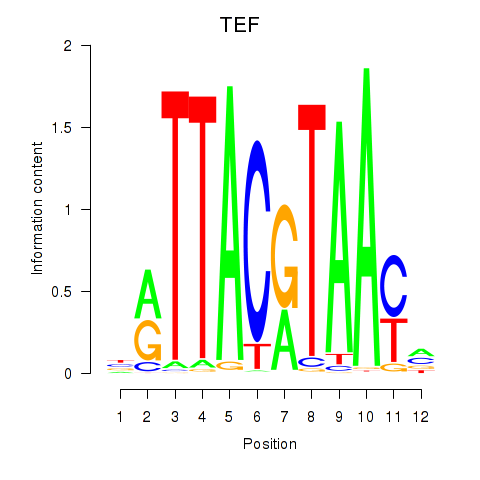
HLF
|
ENSG00000108924.9 | HLF transcription factor, PAR bZIP family member |
|
TEF
|
ENSG00000167074.10 | TEF transcription factor, PAR bZIP family member |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HLF | hg19_v2_chr17_+_53343577_53343588 | 0.32 | 1.2e-01 | Click! |
| TEF | hg19_v2_chr22_+_41777927_41777971, hg19_v2_chr22_+_41763274_41763337 | -0.24 | 2.6e-01 | Click! |
Activity profile of HLF_TEF motif
Sorted Z-values of HLF_TEF motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_27011584 | 2.38 |
ENST00000400532.1
ENST00000480456.1 ENST00000312957.5 |
JAM2
|
junctional adhesion molecule 2 |
| chr1_+_169079823 | 2.03 |
ENST00000367813.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr16_+_70680439 | 1.18 |
ENST00000288098.2
|
IL34
|
interleukin 34 |
| chr1_-_156721389 | 1.12 |
ENST00000537739.1
|
HDGF
|
hepatoma-derived growth factor |
| chr17_-_41623716 | 1.11 |
ENST00000319349.5
|
ETV4
|
ets variant 4 |
| chr17_-_41623009 | 1.11 |
ENST00000393664.2
|
ETV4
|
ets variant 4 |
| chr17_-_41623691 | 1.07 |
ENST00000545954.1
|
ETV4
|
ets variant 4 |
| chr17_-_41623259 | 1.06 |
ENST00000538265.1
ENST00000591713.1 |
ETV4
|
ets variant 4 |
| chr17_-_41623075 | 1.05 |
ENST00000545089.1
|
ETV4
|
ets variant 4 |
| chr7_-_83824169 | 0.90 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr1_+_65613513 | 0.87 |
ENST00000395334.2
|
AK4
|
adenylate kinase 4 |
| chr3_-_190580404 | 0.85 |
ENST00000442080.1
|
GMNC
|
geminin coiled-coil domain containing |
| chr1_-_244013384 | 0.84 |
ENST00000366539.1
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr7_-_22259845 | 0.80 |
ENST00000420196.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr4_-_76928641 | 0.78 |
ENST00000264888.5
|
CXCL9
|
chemokine (C-X-C motif) ligand 9 |
| chr6_-_46459099 | 0.68 |
ENST00000371374.1
|
RCAN2
|
regulator of calcineurin 2 |
| chr1_+_144339738 | 0.67 |
ENST00000538264.1
|
AL592284.1
|
Protein LOC642441 |
| chr7_-_141401951 | 0.67 |
ENST00000536163.1
|
KIAA1147
|
KIAA1147 |
| chr6_+_4706368 | 0.67 |
ENST00000328908.5
|
CDYL
|
chromodomain protein, Y-like |
| chr1_+_7844312 | 0.66 |
ENST00000377541.1
|
PER3
|
period circadian clock 3 |
| chr21_+_45138941 | 0.64 |
ENST00000398081.1
ENST00000468090.1 ENST00000291565.4 |
PDXK
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr14_+_78227105 | 0.61 |
ENST00000439131.2
ENST00000355883.3 ENST00000557011.1 ENST00000556047.1 |
C14orf178
|
chromosome 14 open reading frame 178 |
| chr19_+_41594377 | 0.59 |
ENST00000330436.3
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13 |
| chr11_+_35201826 | 0.58 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr1_+_212738676 | 0.58 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr19_-_41388657 | 0.58 |
ENST00000301146.4
ENST00000291764.3 |
CYP2A7
|
cytochrome P450, family 2, subfamily A, polypeptide 7 |
| chr1_-_156721502 | 0.49 |
ENST00000357325.5
|
HDGF
|
hepatoma-derived growth factor |
| chr4_-_153274078 | 0.49 |
ENST00000263981.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr5_-_54830871 | 0.48 |
ENST00000307259.8
|
PPAP2A
|
phosphatidic acid phosphatase type 2A |
| chr10_-_14372870 | 0.47 |
ENST00000357447.2
|
FRMD4A
|
FERM domain containing 4A |
| chrX_+_9431324 | 0.45 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr3_-_99594948 | 0.42 |
ENST00000471562.1
ENST00000495625.2 |
FILIP1L
|
filamin A interacting protein 1-like |
| chr1_+_174933899 | 0.42 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr2_+_186603355 | 0.41 |
ENST00000343098.5
|
FSIP2
|
fibrous sheath interacting protein 2 |
| chr15_-_83621435 | 0.41 |
ENST00000450735.2
ENST00000426485.1 ENST00000399166.2 ENST00000304231.8 |
HOMER2
|
homer homolog 2 (Drosophila) |
| chr3_+_154797877 | 0.40 |
ENST00000462745.1
ENST00000493237.1 |
MME
|
membrane metallo-endopeptidase |
| chr7_+_90032667 | 0.38 |
ENST00000496677.1
ENST00000287916.4 ENST00000535571.1 ENST00000394604.1 ENST00000394605.2 |
CLDN12
|
claudin 12 |
| chr1_+_144989309 | 0.37 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr18_-_5396271 | 0.36 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr3_+_63953415 | 0.35 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr7_+_23146271 | 0.34 |
ENST00000545771.1
|
KLHL7
|
kelch-like family member 7 |
| chr17_-_41984835 | 0.34 |
ENST00000520406.1
ENST00000518478.1 ENST00000522172.1 ENST00000461854.1 ENST00000521178.1 ENST00000520305.1 ENST00000523501.1 ENST00000520241.1 |
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr3_-_99595037 | 0.33 |
ENST00000383694.2
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr5_+_34757309 | 0.32 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr17_-_39306054 | 0.32 |
ENST00000343246.4
|
KRTAP4-5
|
keratin associated protein 4-5 |
| chr5_-_58882219 | 0.31 |
ENST00000505453.1
ENST00000360047.5 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr7_-_31380502 | 0.31 |
ENST00000297142.3
|
NEUROD6
|
neuronal differentiation 6 |
| chr16_-_54962704 | 0.30 |
ENST00000502066.2
ENST00000560912.1 ENST00000558952.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr21_+_35736302 | 0.30 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chr7_+_23145884 | 0.30 |
ENST00000409689.1
ENST00000410047.1 |
KLHL7
|
kelch-like family member 7 |
| chr11_-_85397167 | 0.29 |
ENST00000316398.3
|
CCDC89
|
coiled-coil domain containing 89 |
| chr9_+_136399929 | 0.28 |
ENST00000393060.1
|
ADAMTSL2
|
ADAMTS-like 2 |
| chr3_-_48130314 | 0.28 |
ENST00000439356.1
ENST00000395734.3 ENST00000426837.2 |
MAP4
|
microtubule-associated protein 4 |
| chr16_-_54962415 | 0.28 |
ENST00000501177.3
ENST00000559598.2 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr22_-_50221160 | 0.27 |
ENST00000404760.1
|
BRD1
|
bromodomain containing 1 |
| chr16_+_56965960 | 0.27 |
ENST00000439977.2
ENST00000344114.4 ENST00000300302.5 ENST00000379792.2 |
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr16_-_54962625 | 0.26 |
ENST00000559432.1
|
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr18_+_21032781 | 0.26 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr15_-_49255632 | 0.26 |
ENST00000332408.4
|
SHC4
|
SHC (Src homology 2 domain containing) family, member 4 |
| chr14_+_96722539 | 0.25 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr12_-_88423164 | 0.25 |
ENST00000298699.2
ENST00000550553.1 |
C12orf50
|
chromosome 12 open reading frame 50 |
| chr19_-_41356347 | 0.24 |
ENST00000301141.5
|
CYP2A6
|
cytochrome P450, family 2, subfamily A, polypeptide 6 |
| chr9_+_71819927 | 0.24 |
ENST00000535702.1
|
TJP2
|
tight junction protein 2 |
| chr9_+_71820057 | 0.24 |
ENST00000539225.1
|
TJP2
|
tight junction protein 2 |
| chr12_-_101801505 | 0.24 |
ENST00000539055.1
ENST00000551688.1 ENST00000551671.1 ENST00000261636.8 |
ARL1
|
ADP-ribosylation factor-like 1 |
| chr3_-_48130707 | 0.23 |
ENST00000360240.6
ENST00000383737.4 |
MAP4
|
microtubule-associated protein 4 |
| chr4_+_96012614 | 0.22 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr16_-_755726 | 0.22 |
ENST00000324361.5
|
FBXL16
|
F-box and leucine-rich repeat protein 16 |
| chr5_+_95998070 | 0.22 |
ENST00000421689.2
ENST00000510756.1 ENST00000512620.1 |
CAST
|
calpastatin |
| chr12_-_53074182 | 0.22 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr17_-_32690239 | 0.22 |
ENST00000225842.3
|
CCL1
|
chemokine (C-C motif) ligand 1 |
| chr16_-_755819 | 0.22 |
ENST00000397621.1
|
FBXL16
|
F-box and leucine-rich repeat protein 16 |
| chr1_+_25757376 | 0.22 |
ENST00000399766.3
ENST00000399763.3 ENST00000374343.4 |
TMEM57
|
transmembrane protein 57 |
| chr11_+_69061594 | 0.22 |
ENST00000441339.2
ENST00000308946.3 ENST00000535407.1 |
MYEOV
|
myeloma overexpressed |
| chr17_+_685513 | 0.21 |
ENST00000304478.4
|
RNMTL1
|
RNA methyltransferase like 1 |
| chr5_-_147162263 | 0.21 |
ENST00000333010.6
ENST00000265272.5 |
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr14_+_60716159 | 0.20 |
ENST00000325658.3
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr3_-_196910721 | 0.20 |
ENST00000443183.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr5_+_95997769 | 0.20 |
ENST00000338252.3
ENST00000508830.1 |
CAST
|
calpastatin |
| chr13_+_23755099 | 0.19 |
ENST00000537476.1
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr12_-_113574028 | 0.19 |
ENST00000546530.1
ENST00000261729.5 |
RASAL1
|
RAS protein activator like 1 (GAP1 like) |
| chr13_-_29292956 | 0.19 |
ENST00000266943.6
|
SLC46A3
|
solute carrier family 46, member 3 |
| chr5_+_172332220 | 0.18 |
ENST00000518247.1
ENST00000326654.2 |
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr9_-_128246769 | 0.18 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr18_+_32621324 | 0.18 |
ENST00000300249.5
ENST00000538170.2 ENST00000588910.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr10_+_94050913 | 0.17 |
ENST00000358935.2
|
MARCH5
|
membrane-associated ring finger (C3HC4) 5 |
| chr14_+_96722152 | 0.17 |
ENST00000216629.6
|
BDKRB1
|
bradykinin receptor B1 |
| chr1_-_247094628 | 0.17 |
ENST00000366508.1
ENST00000326225.3 ENST00000391829.2 |
AHCTF1
|
AT hook containing transcription factor 1 |
| chr14_-_72458326 | 0.17 |
ENST00000542853.1
|
AC005477.1
|
AC005477.1 |
| chr3_+_88188254 | 0.17 |
ENST00000309495.5
|
ZNF654
|
zinc finger protein 654 |
| chr14_+_102276192 | 0.17 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr2_+_198318147 | 0.17 |
ENST00000263960.2
|
COQ10B
|
coenzyme Q10 homolog B (S. cerevisiae) |
| chr2_+_113885138 | 0.16 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr7_+_20687017 | 0.16 |
ENST00000258738.6
|
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr6_+_53948328 | 0.16 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr17_-_61850894 | 0.15 |
ENST00000403162.3
ENST00000582252.1 ENST00000225726.5 |
CCDC47
|
coiled-coil domain containing 47 |
| chr3_+_182983090 | 0.15 |
ENST00000465010.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr9_+_110045537 | 0.14 |
ENST00000358015.3
|
RAD23B
|
RAD23 homolog B (S. cerevisiae) |
| chr5_-_114505624 | 0.14 |
ENST00000513154.1
|
TRIM36
|
tripartite motif containing 36 |
| chr3_+_148447887 | 0.14 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr6_-_64029879 | 0.14 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr2_+_192543153 | 0.14 |
ENST00000425611.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr8_+_50824233 | 0.14 |
ENST00000522124.1
|
SNTG1
|
syntrophin, gamma 1 |
| chr11_-_62313090 | 0.14 |
ENST00000528508.1
ENST00000533365.1 |
AHNAK
|
AHNAK nucleoprotein |
| chr4_-_21950356 | 0.14 |
ENST00000447367.2
ENST00000382152.2 |
KCNIP4
|
Kv channel interacting protein 4 |
| chr2_+_183989157 | 0.13 |
ENST00000541912.1
|
NUP35
|
nucleoporin 35kDa |
| chr8_+_24151553 | 0.13 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr3_-_45883558 | 0.13 |
ENST00000445698.1
ENST00000296135.6 |
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr1_-_178840157 | 0.13 |
ENST00000367629.1
ENST00000234816.2 |
ANGPTL1
|
angiopoietin-like 1 |
| chr14_+_93260569 | 0.13 |
ENST00000163416.2
|
GOLGA5
|
golgin A5 |
| chr1_-_167883327 | 0.13 |
ENST00000476818.2
ENST00000367851.4 ENST00000367848.1 |
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr17_+_61851504 | 0.13 |
ENST00000359353.5
ENST00000389924.2 |
DDX42
|
DEAD (Asp-Glu-Ala-Asp) box helicase 42 |
| chr11_-_18270182 | 0.13 |
ENST00000528349.1
ENST00000526900.1 ENST00000529528.1 ENST00000414546.2 ENST00000256733.4 |
SAA2
|
serum amyloid A2 |
| chr2_-_98280383 | 0.12 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr9_-_114246635 | 0.12 |
ENST00000338205.5
|
KIAA0368
|
KIAA0368 |
| chr1_+_75594119 | 0.12 |
ENST00000294638.5
|
LHX8
|
LIM homeobox 8 |
| chr12_-_54758251 | 0.12 |
ENST00000267015.3
ENST00000551809.1 |
GPR84
|
G protein-coupled receptor 84 |
| chr15_+_49913201 | 0.12 |
ENST00000329873.5
ENST00000558653.1 ENST00000559164.1 ENST00000560632.1 ENST00000559405.1 ENST00000251250.6 |
DTWD1
|
DTW domain containing 1 |
| chr19_+_33865218 | 0.12 |
ENST00000585933.2
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr7_-_45026419 | 0.12 |
ENST00000578968.1
ENST00000580528.1 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chrX_+_36246735 | 0.12 |
ENST00000378653.3
|
CXorf30
|
chromosome X open reading frame 30 |
| chr1_-_167883353 | 0.12 |
ENST00000545172.1
|
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr1_+_204839959 | 0.12 |
ENST00000404076.1
|
NFASC
|
neurofascin |
| chr2_-_31637560 | 0.11 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chr21_+_19617140 | 0.11 |
ENST00000299295.2
ENST00000338326.3 |
CHODL
|
chondrolectin |
| chr11_-_72145426 | 0.11 |
ENST00000535990.1
ENST00000437826.2 ENST00000340729.5 |
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr19_+_49259325 | 0.11 |
ENST00000222157.3
|
FGF21
|
fibroblast growth factor 21 |
| chr2_+_183989083 | 0.11 |
ENST00000295119.4
|
NUP35
|
nucleoporin 35kDa |
| chr17_-_42144949 | 0.11 |
ENST00000591247.1
|
LSM12
|
LSM12 homolog (S. cerevisiae) |
| chr15_+_58430368 | 0.11 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr7_-_45026200 | 0.11 |
ENST00000577700.1
ENST00000580458.1 ENST00000579383.1 ENST00000584686.1 ENST00000585030.1 ENST00000582727.1 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr11_-_77185094 | 0.11 |
ENST00000278568.4
ENST00000356341.3 |
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr1_-_72566613 | 0.11 |
ENST00000306821.3
|
NEGR1
|
neuronal growth regulator 1 |
| chr5_-_146435694 | 0.11 |
ENST00000356826.3
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr7_-_45026159 | 0.11 |
ENST00000584327.1
ENST00000438705.3 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr13_-_33780133 | 0.11 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr9_+_101705893 | 0.11 |
ENST00000375001.3
|
COL15A1
|
collagen, type XV, alpha 1 |
| chr22_-_29457832 | 0.11 |
ENST00000216071.4
|
C22orf31
|
chromosome 22 open reading frame 31 |
| chr2_+_46769798 | 0.11 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr11_-_14993819 | 0.11 |
ENST00000396372.2
ENST00000361010.3 ENST00000359642.3 ENST00000331587.4 |
CALCA
|
calcitonin-related polypeptide alpha |
| chr4_+_41937131 | 0.11 |
ENST00000504986.1
ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33
|
transmembrane protein 33 |
| chr3_-_10547192 | 0.10 |
ENST00000360273.2
ENST00000343816.4 |
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr17_-_19651654 | 0.10 |
ENST00000395555.3
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr4_-_65275162 | 0.10 |
ENST00000381210.3
ENST00000507440.1 |
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr8_+_24151620 | 0.10 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr17_-_39324424 | 0.10 |
ENST00000391356.2
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr16_-_20367584 | 0.10 |
ENST00000570689.1
|
UMOD
|
uromodulin |
| chr15_+_58430567 | 0.10 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr10_-_104211294 | 0.10 |
ENST00000239125.1
|
C10orf95
|
chromosome 10 open reading frame 95 |
| chr19_-_4065730 | 0.10 |
ENST00000601588.1
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr1_+_45212074 | 0.10 |
ENST00000372217.1
|
KIF2C
|
kinesin family member 2C |
| chr5_+_150404904 | 0.09 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr8_+_24298597 | 0.09 |
ENST00000380789.1
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr5_+_95997918 | 0.09 |
ENST00000395812.2
ENST00000395813.1 ENST00000359176.4 ENST00000325674.7 |
CAST
|
calpastatin |
| chr14_-_67826486 | 0.09 |
ENST00000555431.1
ENST00000554236.1 ENST00000555474.1 |
ATP6V1D
|
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
| chr14_+_93260642 | 0.09 |
ENST00000355976.2
|
GOLGA5
|
golgin A5 |
| chr17_-_39538550 | 0.09 |
ENST00000394001.1
|
KRT34
|
keratin 34 |
| chr11_+_62495997 | 0.09 |
ENST00000316461.4
|
TTC9C
|
tetratricopeptide repeat domain 9C |
| chr16_+_3507985 | 0.08 |
ENST00000421765.3
ENST00000360862.5 ENST00000414063.2 ENST00000610180.1 ENST00000608993.1 |
NAA60
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit N-alpha-acetyltransferase 60 |
| chr1_+_45212051 | 0.08 |
ENST00000372222.3
|
KIF2C
|
kinesin family member 2C |
| chr19_+_52076425 | 0.08 |
ENST00000436511.2
|
ZNF175
|
zinc finger protein 175 |
| chr13_-_38172863 | 0.08 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr19_-_35992780 | 0.08 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr8_+_24298531 | 0.08 |
ENST00000175238.6
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr3_+_62304648 | 0.08 |
ENST00000462069.1
ENST00000232519.5 ENST00000465142.1 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chr8_-_102216925 | 0.08 |
ENST00000517844.1
|
ZNF706
|
zinc finger protein 706 |
| chr8_+_77318769 | 0.08 |
ENST00000518732.1
|
RP11-706J10.1
|
long intergenic non-protein coding RNA 1111 |
| chr21_-_30365136 | 0.08 |
ENST00000361371.5
ENST00000389194.2 ENST00000389195.2 |
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chr2_+_103378472 | 0.08 |
ENST00000412401.2
|
TMEM182
|
transmembrane protein 182 |
| chr19_-_3985455 | 0.07 |
ENST00000309311.6
|
EEF2
|
eukaryotic translation elongation factor 2 |
| chr8_+_90914073 | 0.07 |
ENST00000297438.2
|
OSGIN2
|
oxidative stress induced growth inhibitor family member 2 |
| chr17_-_19651668 | 0.07 |
ENST00000494157.2
ENST00000225740.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr6_-_133055896 | 0.07 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr19_+_49258775 | 0.07 |
ENST00000593756.1
|
FGF21
|
fibroblast growth factor 21 |
| chr10_-_82049424 | 0.07 |
ENST00000372213.3
|
MAT1A
|
methionine adenosyltransferase I, alpha |
| chr9_-_123639304 | 0.07 |
ENST00000436309.1
|
PHF19
|
PHD finger protein 19 |
| chr17_-_26941179 | 0.07 |
ENST00000301037.5
ENST00000530121.1 ENST00000525510.1 ENST00000577790.1 ENST00000531839.1 ENST00000534850.1 |
SGK494
RP11-192H23.4
|
uncharacterized serine/threonine-protein kinase SgK494 Uncharacterized protein |
| chr1_-_161207875 | 0.07 |
ENST00000512372.1
ENST00000437437.2 ENST00000442691.2 ENST00000412844.2 ENST00000428574.2 ENST00000505005.1 ENST00000508740.1 ENST00000508387.1 ENST00000504010.1 ENST00000511676.1 ENST00000502985.1 ENST00000367981.3 ENST00000515621.1 ENST00000511944.1 ENST00000511748.1 ENST00000367984.4 ENST00000367985.3 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr14_+_21423611 | 0.07 |
ENST00000304625.2
|
RNASE2
|
ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) |
| chr1_-_161087802 | 0.07 |
ENST00000368010.3
|
PFDN2
|
prefoldin subunit 2 |
| chr1_-_161207953 | 0.07 |
ENST00000367982.4
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr2_-_113594279 | 0.07 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr16_-_30102547 | 0.06 |
ENST00000279386.2
|
TBX6
|
T-box 6 |
| chr2_-_239197201 | 0.06 |
ENST00000254658.3
|
PER2
|
period circadian clock 2 |
| chr19_-_42947121 | 0.06 |
ENST00000601181.1
|
CXCL17
|
chemokine (C-X-C motif) ligand 17 |
| chr10_+_18629628 | 0.06 |
ENST00000377329.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr17_-_685493 | 0.06 |
ENST00000536578.1
ENST00000301328.5 ENST00000576419.1 |
GLOD4
|
glyoxalase domain containing 4 |
| chr1_-_161208013 | 0.06 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr1_-_161207986 | 0.06 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr12_+_57624119 | 0.06 |
ENST00000555773.1
ENST00000554975.1 ENST00000449049.3 ENST00000393827.4 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr9_-_130712995 | 0.06 |
ENST00000373084.4
|
FAM102A
|
family with sequence similarity 102, member A |
| chr1_-_114429997 | 0.06 |
ENST00000471267.1
ENST00000393320.3 |
BCL2L15
|
BCL2-like 15 |
| chr14_+_65453432 | 0.05 |
ENST00000246166.2
|
FNTB
|
farnesyltransferase, CAAX box, beta |
| chr16_+_3508063 | 0.05 |
ENST00000576787.1
ENST00000572942.1 ENST00000576916.1 ENST00000575076.1 ENST00000572131.1 |
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr8_-_102217515 | 0.05 |
ENST00000520347.1
ENST00000523922.1 ENST00000520984.1 |
ZNF706
|
zinc finger protein 706 |
| chr1_+_11249398 | 0.05 |
ENST00000376819.3
|
ANGPTL7
|
angiopoietin-like 7 |
| chr5_+_126988732 | 0.05 |
ENST00000395322.3
|
CTXN3
|
cortexin 3 |
| chr2_-_225362533 | 0.04 |
ENST00000451538.1
|
CUL3
|
cullin 3 |
| chr2_+_102608306 | 0.04 |
ENST00000332549.3
|
IL1R2
|
interleukin 1 receptor, type II |
| chr8_+_7801144 | 0.04 |
ENST00000443676.1
|
ZNF705B
|
zinc finger protein 705B |
Network of associatons between targets according to the STRING database.
First level regulatory network of HLF_TEF
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.2 | 0.6 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.2 | 0.6 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.2 | 0.9 | GO:0048880 | sensory system development(GO:0048880) |
| 0.1 | 0.8 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.1 | 0.5 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.9 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.1 | 0.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.4 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular response to UV-A(GO:0071492) |
| 0.1 | 0.8 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.3 | GO:1902159 | regulation of inward rectifier potassium channel activity(GO:1901979) regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.1 | 0.6 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 1.2 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.1 | 0.3 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.5 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.8 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.5 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.4 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.2 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.5 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.1 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.3 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:1903286 | regulation of potassium ion import(GO:1903286) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:0070487 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) monocyte aggregation(GO:0070487) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.1 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.3 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.7 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.2 | GO:0030952 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) bone regeneration(GO:1990523) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.1 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.4 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.4 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:1900920 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.0 | 0.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.3 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.3 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 2.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.6 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.5 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 3.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 1.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0008478 | pyridoxal kinase activity(GO:0008478) lithium ion binding(GO:0031403) |
| 0.2 | 0.9 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.2 | 0.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 1.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.4 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 2.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 1.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.4 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.5 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.8 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.3 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.2 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 0.2 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.3 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 1.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 5.8 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.8 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.6 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.8 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.9 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 2.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.8 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 1.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 2.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |