Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
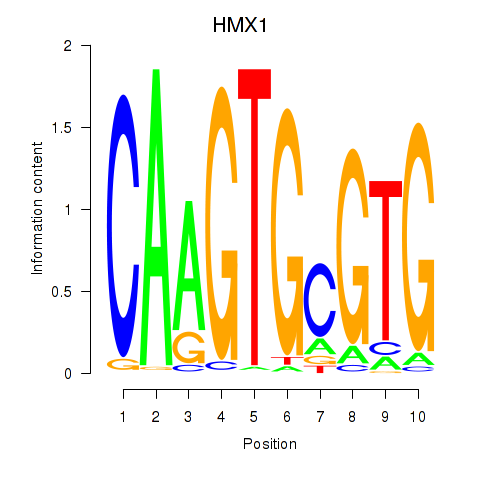
Results for HMX1
Z-value: 0.80
Transcription factors associated with HMX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMX1
|
ENSG00000215612.5 | H6 family homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMX1 | hg19_v2_chr4_-_8873531_8873543 | 0.04 | 8.5e-01 | Click! |
Activity profile of HMX1 motif
Sorted Z-values of HMX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_49259643 | 1.89 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr12_+_57482665 | 1.29 |
ENST00000300131.3
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr5_-_81046904 | 1.20 |
ENST00000515395.1
|
SSBP2
|
single-stranded DNA binding protein 2 |
| chr12_+_57482877 | 1.20 |
ENST00000342556.6
ENST00000357680.4 |
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr5_-_81046841 | 1.15 |
ENST00000509013.2
ENST00000505980.1 ENST00000509053.1 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr4_-_186733363 | 1.14 |
ENST00000393523.2
ENST00000393528.3 ENST00000449407.2 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr11_+_59522900 | 1.12 |
ENST00000529177.1
|
STX3
|
syntaxin 3 |
| chr19_+_1407733 | 1.09 |
ENST00000592453.1
|
DAZAP1
|
DAZ associated protein 1 |
| chr14_+_63671577 | 0.96 |
ENST00000555125.1
|
RHOJ
|
ras homolog family member J |
| chr11_+_59522837 | 0.95 |
ENST00000437946.2
|
STX3
|
syntaxin 3 |
| chr16_+_88872176 | 0.92 |
ENST00000569140.1
|
CDT1
|
chromatin licensing and DNA replication factor 1 |
| chr9_-_130617029 | 0.71 |
ENST00000373203.4
|
ENG
|
endoglin |
| chr1_-_41328018 | 0.70 |
ENST00000372638.2
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr9_-_130616915 | 0.69 |
ENST00000344849.3
|
ENG
|
endoglin |
| chr2_+_106682135 | 0.68 |
ENST00000437659.1
|
C2orf40
|
chromosome 2 open reading frame 40 |
| chr2_+_106682103 | 0.66 |
ENST00000238044.3
|
C2orf40
|
chromosome 2 open reading frame 40 |
| chr7_-_156685841 | 0.66 |
ENST00000354505.4
ENST00000540390.1 |
LMBR1
|
limb development membrane protein 1 |
| chr12_+_53440753 | 0.63 |
ENST00000379902.3
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr9_+_82186682 | 0.60 |
ENST00000376552.2
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr18_-_45935663 | 0.60 |
ENST00000589194.1
ENST00000591279.1 ENST00000590855.1 ENST00000587107.1 ENST00000588970.1 ENST00000586525.1 ENST00000592387.1 ENST00000590800.1 |
ZBTB7C
|
zinc finger and BTB domain containing 7C |
| chr17_+_66509019 | 0.59 |
ENST00000585981.1
ENST00000589480.1 ENST00000585815.1 |
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr11_-_128392085 | 0.57 |
ENST00000526145.2
ENST00000531611.1 ENST00000319397.6 ENST00000345075.4 ENST00000535549.1 |
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr15_+_89402148 | 0.57 |
ENST00000560601.1
|
ACAN
|
aggrecan |
| chr14_+_56585048 | 0.56 |
ENST00000267460.4
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr11_-_64815333 | 0.56 |
ENST00000533842.1
ENST00000532802.1 ENST00000530139.1 ENST00000526516.1 |
NAALADL1
|
N-acetylated alpha-linked acidic dipeptidase-like 1 |
| chr3_+_5020801 | 0.54 |
ENST00000256495.3
|
BHLHE40
|
basic helix-loop-helix family, member e40 |
| chr1_-_27286897 | 0.51 |
ENST00000320567.5
|
C1orf172
|
chromosome 1 open reading frame 172 |
| chr21_-_40032581 | 0.50 |
ENST00000398919.2
|
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr9_+_82186872 | 0.49 |
ENST00000376544.3
ENST00000376520.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chrX_+_53123314 | 0.48 |
ENST00000605526.1
ENST00000604062.1 ENST00000604369.1 ENST00000366185.2 ENST00000604849.1 |
RP11-258C19.5
|
long intergenic non-protein coding RNA 1155 |
| chr5_+_139027877 | 0.46 |
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5 |
| chr3_+_20081515 | 0.45 |
ENST00000263754.4
|
KAT2B
|
K(lysine) acetyltransferase 2B |
| chr4_+_86396321 | 0.43 |
ENST00000503995.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr17_-_6947225 | 0.43 |
ENST00000574600.1
ENST00000308009.1 ENST00000447225.1 |
SLC16A11
|
solute carrier family 16, member 11 |
| chr3_-_128294929 | 0.42 |
ENST00000356020.2
|
C3orf27
|
chromosome 3 open reading frame 27 |
| chr17_+_79989937 | 0.42 |
ENST00000580965.1
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chr1_-_85930246 | 0.42 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr13_+_25254545 | 0.39 |
ENST00000218548.6
|
ATP12A
|
ATPase, H+/K+ transporting, nongastric, alpha polypeptide |
| chr22_-_18923655 | 0.38 |
ENST00000438924.1
ENST00000457083.1 ENST00000420436.1 ENST00000334029.2 ENST00000357068.6 |
PRODH
|
proline dehydrogenase (oxidase) 1 |
| chr9_-_140082983 | 0.38 |
ENST00000323927.2
|
ANAPC2
|
anaphase promoting complex subunit 2 |
| chr3_+_101546827 | 0.37 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr3_+_127391769 | 0.37 |
ENST00000393363.3
ENST00000232744.8 ENST00000453791.2 |
ABTB1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr1_-_167059830 | 0.37 |
ENST00000367868.3
|
GPA33
|
glycoprotein A33 (transmembrane) |
| chr12_+_53400176 | 0.37 |
ENST00000551002.1
ENST00000420463.3 ENST00000416762.3 ENST00000549481.1 ENST00000552490.1 |
EIF4B
|
eukaryotic translation initiation factor 4B |
| chr19_-_46526304 | 0.36 |
ENST00000008938.4
|
PGLYRP1
|
peptidoglycan recognition protein 1 |
| chr17_+_79989500 | 0.36 |
ENST00000306897.4
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chr3_+_151986709 | 0.35 |
ENST00000495875.2
ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr4_-_80993717 | 0.35 |
ENST00000307333.7
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr17_-_7165662 | 0.34 |
ENST00000571881.2
ENST00000360325.7 |
CLDN7
|
claudin 7 |
| chrX_+_13587712 | 0.34 |
ENST00000361306.1
ENST00000380602.3 |
EGFL6
|
EGF-like-domain, multiple 6 |
| chr20_-_35374456 | 0.34 |
ENST00000373803.2
ENST00000359675.2 ENST00000540765.1 ENST00000349004.1 |
NDRG3
|
NDRG family member 3 |
| chr3_+_57994127 | 0.32 |
ENST00000490882.1
ENST00000295956.4 ENST00000358537.3 ENST00000429972.2 ENST00000348383.5 ENST00000357272.4 |
FLNB
|
filamin B, beta |
| chr17_-_74533734 | 0.32 |
ENST00000589342.1
|
CYGB
|
cytoglobin |
| chr5_+_156693091 | 0.31 |
ENST00000318218.6
ENST00000442283.2 ENST00000522463.1 ENST00000521420.1 |
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr1_-_53387386 | 0.31 |
ENST00000467988.1
ENST00000358358.5 ENST00000371522.4 |
ECHDC2
|
enoyl CoA hydratase domain containing 2 |
| chr17_+_79990058 | 0.31 |
ENST00000584341.1
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chr10_-_99531709 | 0.30 |
ENST00000266066.3
|
SFRP5
|
secreted frizzled-related protein 5 |
| chr9_-_131644202 | 0.30 |
ENST00000320665.6
ENST00000436267.2 |
CCBL1
|
cysteine conjugate-beta lyase, cytoplasmic |
| chr1_-_154946825 | 0.30 |
ENST00000368453.4
ENST00000368450.1 ENST00000366442.2 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr17_-_62097904 | 0.29 |
ENST00000583366.1
|
ICAM2
|
intercellular adhesion molecule 2 |
| chr17_-_73874654 | 0.29 |
ENST00000254816.2
|
TRIM47
|
tripartite motif containing 47 |
| chr22_-_37640456 | 0.29 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr4_-_11430221 | 0.29 |
ENST00000514690.1
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr19_-_14316980 | 0.28 |
ENST00000361434.3
ENST00000340736.6 |
LPHN1
|
latrophilin 1 |
| chrX_+_70443050 | 0.28 |
ENST00000361726.6
|
GJB1
|
gap junction protein, beta 1, 32kDa |
| chr1_-_46598371 | 0.28 |
ENST00000372006.1
ENST00000425892.1 ENST00000420542.1 ENST00000354242.4 ENST00000340332.6 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr1_-_32229523 | 0.27 |
ENST00000398547.1
ENST00000373655.2 ENST00000373658.3 ENST00000257070.4 |
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr18_+_11981427 | 0.27 |
ENST00000269159.3
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr12_+_53399942 | 0.27 |
ENST00000262056.9
|
EIF4B
|
eukaryotic translation initiation factor 4B |
| chr19_-_1401486 | 0.27 |
ENST00000252288.2
ENST00000447102.3 |
GAMT
|
guanidinoacetate N-methyltransferase |
| chr7_-_156685890 | 0.27 |
ENST00000353442.5
|
LMBR1
|
limb development membrane protein 1 |
| chr8_+_145691411 | 0.27 |
ENST00000301332.2
|
KIFC2
|
kinesin family member C2 |
| chr4_+_86396265 | 0.26 |
ENST00000395184.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr13_+_25254693 | 0.26 |
ENST00000381946.3
|
ATP12A
|
ATPase, H+/K+ transporting, nongastric, alpha polypeptide |
| chr2_+_204801471 | 0.26 |
ENST00000316386.6
ENST00000435193.1 |
ICOS
|
inducible T-cell co-stimulator |
| chr5_+_156693159 | 0.26 |
ENST00000347377.6
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr1_-_46598284 | 0.26 |
ENST00000423209.1
ENST00000262741.5 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr1_-_32229934 | 0.26 |
ENST00000398542.1
|
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr3_+_49711777 | 0.26 |
ENST00000442186.1
ENST00000438011.1 ENST00000457042.1 |
APEH
|
acylaminoacyl-peptide hydrolase |
| chr8_-_105479270 | 0.25 |
ENST00000521573.2
ENST00000351513.2 |
DPYS
|
dihydropyrimidinase |
| chr7_+_155250824 | 0.25 |
ENST00000297375.4
|
EN2
|
engrailed homeobox 2 |
| chr22_+_47070490 | 0.25 |
ENST00000408031.1
|
GRAMD4
|
GRAM domain containing 4 |
| chr7_-_132261253 | 0.25 |
ENST00000321063.4
|
PLXNA4
|
plexin A4 |
| chr1_+_39546988 | 0.24 |
ENST00000484793.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr1_-_33168336 | 0.24 |
ENST00000373484.3
|
SYNC
|
syncoilin, intermediate filament protein |
| chr22_-_38349552 | 0.23 |
ENST00000422191.1
ENST00000249079.2 ENST00000418863.1 ENST00000403305.1 ENST00000403026.1 |
C22orf23
|
chromosome 22 open reading frame 23 |
| chr17_-_15244894 | 0.23 |
ENST00000338696.2
ENST00000543896.1 ENST00000539245.1 ENST00000539316.1 ENST00000395930.1 |
TEKT3
|
tektin 3 |
| chr12_-_55378452 | 0.23 |
ENST00000449076.1
|
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr19_-_10697895 | 0.23 |
ENST00000591240.1
ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr15_+_24920541 | 0.23 |
ENST00000329468.2
|
NPAP1
|
nuclear pore associated protein 1 |
| chr5_-_81046922 | 0.23 |
ENST00000514493.1
ENST00000320672.4 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr17_-_62097927 | 0.22 |
ENST00000578313.1
ENST00000584084.1 ENST00000579788.1 ENST00000579687.1 ENST00000578379.1 ENST00000578892.1 ENST00000412356.1 ENST00000418105.1 |
ICAM2
|
intercellular adhesion molecule 2 |
| chr6_+_41606176 | 0.22 |
ENST00000441667.1
ENST00000230321.6 ENST00000373050.4 ENST00000446650.1 ENST00000435476.1 |
MDFI
|
MyoD family inhibitor |
| chr17_+_77751931 | 0.22 |
ENST00000310942.4
ENST00000269399.5 |
CBX2
|
chromobox homolog 2 |
| chr9_-_131644306 | 0.22 |
ENST00000302586.3
|
CCBL1
|
cysteine conjugate-beta lyase, cytoplasmic |
| chr14_-_25103388 | 0.22 |
ENST00000526004.1
ENST00000415355.3 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr7_+_31009148 | 0.22 |
ENST00000409904.3
ENST00000409316.1 |
GHRHR
|
growth hormone releasing hormone receptor |
| chr9_+_10613163 | 0.22 |
ENST00000429581.2
|
RP11-87N24.2
|
RP11-87N24.2 |
| chr21_-_40685536 | 0.21 |
ENST00000341322.4
|
BRWD1
|
bromodomain and WD repeat domain containing 1 |
| chr14_-_25103472 | 0.21 |
ENST00000216341.4
ENST00000382542.1 ENST00000382540.1 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr14_+_63671105 | 0.21 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr2_-_27545921 | 0.21 |
ENST00000402310.1
ENST00000405983.1 ENST00000403262.2 ENST00000428910.1 ENST00000402722.1 ENST00000399052.4 ENST00000380044.1 ENST00000405076.1 |
MPV17
|
MpV17 mitochondrial inner membrane protein |
| chr7_-_37488834 | 0.21 |
ENST00000310758.4
|
ELMO1
|
engulfment and cell motility 1 |
| chr3_+_39093481 | 0.21 |
ENST00000302313.5
ENST00000544962.1 ENST00000396258.3 ENST00000418020.1 |
WDR48
|
WD repeat domain 48 |
| chr11_-_417388 | 0.21 |
ENST00000332725.3
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr12_+_41831485 | 0.21 |
ENST00000539469.2
ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr3_+_69788576 | 0.21 |
ENST00000352241.4
ENST00000448226.2 |
MITF
|
microphthalmia-associated transcription factor |
| chr9_+_131644398 | 0.20 |
ENST00000372599.3
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr20_+_30640004 | 0.20 |
ENST00000520553.1
ENST00000518730.1 ENST00000375852.2 |
HCK
|
hemopoietic cell kinase |
| chr3_-_107941209 | 0.20 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr2_-_27712583 | 0.20 |
ENST00000260570.3
ENST00000359466.6 ENST00000416524.2 |
IFT172
|
intraflagellar transport 172 homolog (Chlamydomonas) |
| chr1_+_39547082 | 0.20 |
ENST00000602421.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr11_-_417308 | 0.20 |
ENST00000397632.3
ENST00000382520.2 |
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr1_-_51425902 | 0.20 |
ENST00000396153.2
|
FAF1
|
Fas (TNFRSF6) associated factor 1 |
| chr3_+_50192537 | 0.20 |
ENST00000002829.3
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr3_-_125775629 | 0.20 |
ENST00000383598.2
|
SLC41A3
|
solute carrier family 41, member 3 |
| chr3_+_50192499 | 0.19 |
ENST00000413852.1
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr20_+_30639991 | 0.19 |
ENST00000534862.1
ENST00000538448.1 ENST00000375862.2 |
HCK
|
hemopoietic cell kinase |
| chr2_-_27545431 | 0.19 |
ENST00000233545.2
|
MPV17
|
MpV17 mitochondrial inner membrane protein |
| chr9_+_131644388 | 0.19 |
ENST00000372600.4
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr11_-_33743952 | 0.19 |
ENST00000534312.1
|
CD59
|
CD59 molecule, complement regulatory protein |
| chr12_+_81110684 | 0.19 |
ENST00000228644.3
|
MYF5
|
myogenic factor 5 |
| chr17_+_79679369 | 0.19 |
ENST00000350690.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr2_+_10091783 | 0.19 |
ENST00000324883.5
|
GRHL1
|
grainyhead-like 1 (Drosophila) |
| chr2_-_197457335 | 0.19 |
ENST00000260983.3
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr17_+_79679299 | 0.19 |
ENST00000331531.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr18_+_11981547 | 0.18 |
ENST00000588927.1
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr2_+_10091815 | 0.18 |
ENST00000324907.9
|
GRHL1
|
grainyhead-like 1 (Drosophila) |
| chr6_-_2245892 | 0.18 |
ENST00000380815.4
|
GMDS
|
GDP-mannose 4,6-dehydratase |
| chr3_-_52864680 | 0.18 |
ENST00000406595.1
ENST00000485816.1 ENST00000434759.3 ENST00000346281.5 ENST00000266041.4 |
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr9_+_131644781 | 0.18 |
ENST00000259324.5
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chrX_-_20284958 | 0.17 |
ENST00000379565.3
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr20_+_3451732 | 0.17 |
ENST00000446916.2
|
ATRN
|
attractin |
| chr19_-_2308127 | 0.17 |
ENST00000404279.1
|
LINGO3
|
leucine rich repeat and Ig domain containing 3 |
| chrX_+_110924346 | 0.17 |
ENST00000371979.3
ENST00000251943.4 ENST00000486353.1 ENST00000394780.3 ENST00000495283.1 |
ALG13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr5_-_172662303 | 0.16 |
ENST00000517440.1
ENST00000329198.4 |
NKX2-5
|
NK2 homeobox 5 |
| chr11_-_116968987 | 0.16 |
ENST00000434315.2
ENST00000292055.4 ENST00000375288.1 ENST00000542607.1 ENST00000445177.1 ENST00000375300.1 ENST00000446921.2 |
SIK3
|
SIK family kinase 3 |
| chr8_+_145438870 | 0.16 |
ENST00000527931.1
|
FAM203B
|
family with sequence similarity 203, member B |
| chr16_+_3062457 | 0.16 |
ENST00000445369.2
|
CLDN9
|
claudin 9 |
| chr1_+_160336851 | 0.16 |
ENST00000302101.5
|
NHLH1
|
nescient helix loop helix 1 |
| chr7_+_130131907 | 0.16 |
ENST00000223215.4
ENST00000437945.1 |
MEST
|
mesoderm specific transcript |
| chr4_-_141348999 | 0.16 |
ENST00000325617.5
|
CLGN
|
calmegin |
| chr19_-_19006920 | 0.16 |
ENST00000429504.2
ENST00000427170.2 |
CERS1
|
ceramide synthase 1 |
| chr10_+_99258625 | 0.16 |
ENST00000370664.3
|
UBTD1
|
ubiquitin domain containing 1 |
| chr19_+_41305330 | 0.16 |
ENST00000593972.1
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr4_-_141348789 | 0.15 |
ENST00000414773.1
|
CLGN
|
calmegin |
| chr8_-_67525473 | 0.15 |
ENST00000522677.3
|
MYBL1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr8_+_32406179 | 0.15 |
ENST00000405005.3
|
NRG1
|
neuregulin 1 |
| chr3_-_138553779 | 0.15 |
ENST00000461451.1
|
PIK3CB
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr12_+_70132632 | 0.15 |
ENST00000378815.6
ENST00000483530.2 ENST00000325555.9 |
RAB3IP
|
RAB3A interacting protein |
| chr17_-_72709050 | 0.15 |
ENST00000583937.1
ENST00000301573.9 ENST00000326165.6 ENST00000469092.1 |
CD300LF
|
CD300 molecule-like family member f |
| chr6_+_30131318 | 0.15 |
ENST00000376688.1
|
TRIM15
|
tripartite motif containing 15 |
| chr9_-_130331297 | 0.15 |
ENST00000373312.3
|
FAM129B
|
family with sequence similarity 129, member B |
| chr10_+_126150369 | 0.15 |
ENST00000392757.4
ENST00000368842.5 ENST00000368839.1 |
LHPP
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr22_+_38349670 | 0.15 |
ENST00000442738.2
ENST00000460648.1 ENST00000407936.1 ENST00000488684.1 ENST00000492213.1 ENST00000606538.1 ENST00000405557.1 |
POLR2F
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr19_-_10687907 | 0.14 |
ENST00000589348.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr10_-_135122603 | 0.14 |
ENST00000368563.2
|
TUBGCP2
|
tubulin, gamma complex associated protein 2 |
| chr9_+_137967366 | 0.14 |
ENST00000252854.4
|
OLFM1
|
olfactomedin 1 |
| chr3_-_88198965 | 0.14 |
ENST00000467332.1
ENST00000462901.1 |
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr1_-_51425772 | 0.14 |
ENST00000371778.4
|
FAF1
|
Fas (TNFRSF6) associated factor 1 |
| chr1_-_24194771 | 0.14 |
ENST00000374479.3
|
FUCA1
|
fucosidase, alpha-L- 1, tissue |
| chr9_-_116061476 | 0.14 |
ENST00000441031.3
|
RNF183
|
ring finger protein 183 |
| chr19_-_7939319 | 0.14 |
ENST00000539422.1
|
CTD-3193O13.9
|
Protein FLJ22184 |
| chr6_+_2246023 | 0.14 |
ENST00000530833.1
ENST00000525811.1 ENST00000534441.1 ENST00000533653.1 ENST00000534468.1 |
GMDS-AS1
|
GMDS antisense RNA 1 (head to head) |
| chr6_-_8102714 | 0.14 |
ENST00000502429.1
ENST00000429723.2 ENST00000507463.1 ENST00000379715.5 |
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr5_+_141016969 | 0.14 |
ENST00000518856.1
|
RELL2
|
RELT-like 2 |
| chr1_-_207095212 | 0.14 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr14_+_23564700 | 0.13 |
ENST00000554203.1
|
C14orf119
|
chromosome 14 open reading frame 119 |
| chr5_+_141016508 | 0.13 |
ENST00000444782.1
ENST00000521367.1 ENST00000297164.3 |
RELL2
|
RELT-like 2 |
| chr8_+_87878640 | 0.13 |
ENST00000518476.1
|
CNBD1
|
cyclic nucleotide binding domain containing 1 |
| chr17_+_66508537 | 0.13 |
ENST00000392711.1
ENST00000585427.1 ENST00000589228.1 ENST00000536854.2 ENST00000588702.1 ENST00000589309.1 |
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr1_+_154947126 | 0.13 |
ENST00000368439.1
|
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr16_+_56623433 | 0.13 |
ENST00000570176.1
|
MT3
|
metallothionein 3 |
| chr20_+_21284416 | 0.13 |
ENST00000539513.1
|
XRN2
|
5'-3' exoribonuclease 2 |
| chr1_-_109940550 | 0.13 |
ENST00000256637.6
|
SORT1
|
sortilin 1 |
| chr1_+_154947148 | 0.13 |
ENST00000368436.1
ENST00000308987.5 |
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr6_+_2245982 | 0.13 |
ENST00000530346.1
ENST00000524770.1 ENST00000532124.1 ENST00000531092.1 ENST00000456943.2 ENST00000529893.1 |
GMDS-AS1
|
GMDS antisense RNA 1 (head to head) |
| chr13_+_35516390 | 0.13 |
ENST00000540320.1
ENST00000400445.3 ENST00000310336.4 |
NBEA
|
neurobeachin |
| chr1_-_17338267 | 0.13 |
ENST00000326735.8
|
ATP13A2
|
ATPase type 13A2 |
| chr3_-_49066811 | 0.12 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chrX_-_153285251 | 0.12 |
ENST00000444230.1
ENST00000393682.1 ENST00000393687.2 ENST00000429936.2 ENST00000369974.2 |
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr11_-_110561721 | 0.12 |
ENST00000357139.3
|
ARHGAP20
|
Rho GTPase activating protein 20 |
| chr3_-_18466787 | 0.12 |
ENST00000338745.6
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr19_+_41305612 | 0.12 |
ENST00000594380.1
ENST00000593397.1 ENST00000601733.1 |
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr12_-_7077125 | 0.12 |
ENST00000545555.2
|
PHB2
|
prohibitin 2 |
| chr20_+_21283941 | 0.12 |
ENST00000377191.3
ENST00000430571.2 |
XRN2
|
5'-3' exoribonuclease 2 |
| chr9_+_116263639 | 0.12 |
ENST00000343817.5
|
RGS3
|
regulator of G-protein signaling 3 |
| chr11_-_33744256 | 0.12 |
ENST00000415002.2
ENST00000437761.2 ENST00000445143.2 |
CD59
|
CD59 molecule, complement regulatory protein |
| chr7_-_752577 | 0.12 |
ENST00000544935.1
ENST00000430040.1 ENST00000456696.2 ENST00000406797.1 |
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr16_-_188624 | 0.11 |
ENST00000399953.3
|
NPRL3
|
nitrogen permease regulator-like 3 (S. cerevisiae) |
| chr20_+_49348081 | 0.11 |
ENST00000371610.2
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr19_+_41305627 | 0.11 |
ENST00000593525.1
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr16_+_12995614 | 0.11 |
ENST00000423335.2
|
SHISA9
|
shisa family member 9 |
| chr22_+_47158578 | 0.11 |
ENST00000355704.3
|
TBC1D22A
|
TBC1 domain family, member 22A |
| chr1_-_110052302 | 0.11 |
ENST00000369864.4
ENST00000369862.1 |
AMIGO1
|
adhesion molecule with Ig-like domain 1 |
| chrX_+_135579238 | 0.11 |
ENST00000535601.1
ENST00000448450.1 ENST00000425695.1 |
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr5_+_10353780 | 0.11 |
ENST00000449913.2
ENST00000503788.1 ENST00000274140.5 |
MARCH6
|
membrane-associated ring finger (C3HC4) 6, E3 ubiquitin protein ligase |
| chr9_-_116061145 | 0.11 |
ENST00000416588.2
|
RNF183
|
ring finger protein 183 |
| chr9_+_125273081 | 0.11 |
ENST00000335302.5
|
OR1J2
|
olfactory receptor, family 1, subfamily J, member 2 |
| chr15_-_41166414 | 0.11 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
Network of associatons between targets according to the STRING database.
First level regulatory network of HMX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.4 | 2.5 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.3 | 0.9 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.3 | 2.1 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 0.6 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.4 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.1 | 0.4 | GO:0036484 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.1 | 0.4 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.1 | 0.4 | GO:0051714 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 1.9 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.5 | GO:0035948 | N-terminal peptidyl-lysine acetylation(GO:0018076) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 1.2 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.5 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.5 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 1.1 | GO:0014041 | regulation of neuron maturation(GO:0014041) |
| 0.1 | 0.2 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 0.3 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.1 | 0.4 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.1 | 0.5 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 0.3 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.1 | 1.3 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 0.2 | GO:1905166 | negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.1 | 0.6 | GO:0030578 | PML body organization(GO:0030578) |
| 0.1 | 0.6 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 0.3 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) beta-alanine metabolic process(GO:0019482) thymine metabolic process(GO:0019859) uracil metabolic process(GO:0019860) |
| 0.1 | 0.4 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 0.2 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.1 | 0.4 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.2 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.2 | GO:0003285 | atrioventricular node development(GO:0003162) right ventricular cardiac muscle tissue morphogenesis(GO:0003221) septum secundum development(GO:0003285) |
| 0.1 | 0.2 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 0.2 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.1 | 0.2 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.1 | 0.3 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.1 | GO:0034125 | negative regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034125) regulation of interleukin-4-mediated signaling pathway(GO:1902214) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.5 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:2000374 | cadmium ion homeostasis(GO:0055073) lysosomal membrane organization(GO:0097212) negative regulation of hydrogen peroxide catabolic process(GO:2000296) regulation of oxygen metabolic process(GO:2000374) |
| 0.0 | 0.6 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.1 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.0 | 1.1 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 1.0 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.4 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.4 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 0.6 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.2 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.3 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.3 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.2 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.2 | GO:0061525 | hindgut development(GO:0061525) |
| 0.0 | 0.0 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 1.0 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.4 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.5 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.1 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.8 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.3 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.3 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.2 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.8 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.2 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.2 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.2 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.5 | 1.4 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.2 | 0.6 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.1 | 0.2 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.1 | 0.2 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.1 | 0.4 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 0.5 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.3 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.1 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 0.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.2 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.5 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.2 | 1.4 | GO:0005534 | galactose binding(GO:0005534) |
| 0.2 | 0.6 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.2 | 0.6 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.4 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.1 | 0.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.5 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.4 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.1 | 0.4 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.2 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.3 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.0 | 1.0 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 1.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.2 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.3 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 2.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.0 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 1.0 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.0 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.7 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 2.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.7 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.9 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.8 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 1.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 3.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |