Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for HMX3
Z-value: 0.87
Transcription factors associated with HMX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMX3
|
ENSG00000188620.9 | H6 family homeobox 3 |
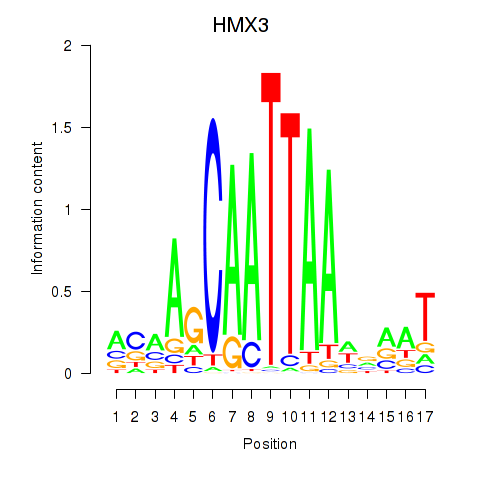
Activity profile of HMX3 motif
Sorted Z-values of HMX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_101439242 | 2.96 |
ENST00000296420.4
|
EMCN
|
endomucin |
| chr4_+_156588249 | 2.79 |
ENST00000393832.3
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr4_-_90756769 | 2.48 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr4_-_101439148 | 2.32 |
ENST00000511970.1
ENST00000502569.1 ENST00000305864.3 |
EMCN
|
endomucin |
| chr12_-_10022735 | 2.17 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr17_+_39394250 | 2.07 |
ENST00000254072.6
|
KRTAP9-8
|
keratin associated protein 9-8 |
| chr4_-_90759440 | 2.00 |
ENST00000336904.3
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr2_+_173686303 | 1.81 |
ENST00000397087.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr9_+_71986182 | 1.77 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr8_-_93029865 | 1.73 |
ENST00000422361.2
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr19_-_41985048 | 1.72 |
ENST00000599801.1
ENST00000595063.1 ENST00000598215.1 |
AC011526.1
|
AC011526.1 |
| chr4_-_90757364 | 1.60 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr4_-_159080806 | 1.55 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr12_+_51318513 | 1.50 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr2_+_89890533 | 1.49 |
ENST00000429992.2
|
IGKV2D-40
|
immunoglobulin kappa variable 2D-40 |
| chr10_-_7513904 | 1.46 |
ENST00000420395.1
|
RP5-1031D4.2
|
RP5-1031D4.2 |
| chr6_-_76203454 | 1.41 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chrX_-_154033661 | 1.40 |
ENST00000393531.1
|
MPP1
|
membrane protein, palmitoylated 1, 55kDa |
| chr1_-_110933611 | 1.20 |
ENST00000472422.2
ENST00000437429.2 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr2_-_191115229 | 1.18 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr21_+_17909594 | 1.15 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr5_-_55412774 | 1.14 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr10_-_126849588 | 1.11 |
ENST00000411419.2
|
CTBP2
|
C-terminal binding protein 2 |
| chr7_+_150413645 | 1.11 |
ENST00000307194.5
|
GIMAP1
|
GTPase, IMAP family member 1 |
| chr5_+_140227048 | 1.11 |
ENST00000532602.1
|
PCDHA9
|
protocadherin alpha 9 |
| chr1_+_109102652 | 1.08 |
ENST00000370035.3
ENST00000405454.1 |
FAM102B
|
family with sequence similarity 102, member B |
| chr6_-_84419101 | 1.08 |
ENST00000520302.1
ENST00000520213.1 ENST00000439399.2 ENST00000428679.2 ENST00000437520.1 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr14_-_105420241 | 1.07 |
ENST00000557457.1
|
AHNAK2
|
AHNAK nucleoprotein 2 |
| chr9_+_99690592 | 1.07 |
ENST00000354649.3
|
NUTM2G
|
NUT family member 2G |
| chr16_+_20775358 | 1.03 |
ENST00000440284.2
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr2_-_136873735 | 1.01 |
ENST00000409817.1
|
CXCR4
|
chemokine (C-X-C motif) receptor 4 |
| chr8_+_97506033 | 1.00 |
ENST00000518385.1
|
SDC2
|
syndecan 2 |
| chr5_+_112849373 | 0.98 |
ENST00000161863.4
ENST00000515883.1 |
YTHDC2
|
YTH domain containing 2 |
| chr6_-_52859046 | 0.97 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr5_+_121647386 | 0.97 |
ENST00000542191.1
ENST00000506272.1 ENST00000508681.1 ENST00000509154.2 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr4_-_104021009 | 0.97 |
ENST00000509245.1
ENST00000296424.4 |
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr14_+_64680854 | 0.96 |
ENST00000458046.2
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr4_+_90823130 | 0.94 |
ENST00000508372.1
|
MMRN1
|
multimerin 1 |
| chr15_+_25200074 | 0.92 |
ENST00000390687.4
ENST00000584968.1 ENST00000346403.6 ENST00000554227.2 |
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chr11_+_62104897 | 0.91 |
ENST00000415229.2
ENST00000535727.1 ENST00000301776.5 |
ASRGL1
|
asparaginase like 1 |
| chr17_-_295730 | 0.90 |
ENST00000329099.4
|
FAM101B
|
family with sequence similarity 101, member B |
| chr11_+_7618413 | 0.89 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr1_+_76540386 | 0.89 |
ENST00000328299.3
|
ST6GALNAC3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr6_+_168196210 | 0.87 |
ENST00000597278.1
|
AL009178.1
|
Uncharacterized protein; cDNA FLJ43200 fis, clone FEBRA2007793 |
| chr22_-_29107919 | 0.87 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr1_-_21377447 | 0.86 |
ENST00000374937.3
ENST00000264211.8 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr1_-_110933663 | 0.85 |
ENST00000369781.4
ENST00000541986.1 ENST00000369779.4 |
SLC16A4
|
solute carrier family 16, member 4 |
| chrX_+_13671225 | 0.84 |
ENST00000545566.1
ENST00000544987.1 ENST00000314720.4 |
TCEANC
|
transcription elongation factor A (SII) N-terminal and central domain containing |
| chr17_+_32597232 | 0.83 |
ENST00000378569.2
ENST00000200307.4 ENST00000394627.1 ENST00000394630.3 |
CCL7
|
chemokine (C-C motif) ligand 7 |
| chr12_-_27167233 | 0.81 |
ENST00000535819.1
ENST00000543803.1 ENST00000535423.1 ENST00000539741.1 ENST00000343028.4 ENST00000545600.1 ENST00000543088.1 |
TM7SF3
|
transmembrane 7 superfamily member 3 |
| chr12_-_6055398 | 0.81 |
ENST00000327087.8
ENST00000356134.5 ENST00000546188.1 |
ANO2
|
anoctamin 2 |
| chr5_-_94417339 | 0.80 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr8_-_17555164 | 0.80 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr17_+_67498538 | 0.79 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr5_+_95066823 | 0.79 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr11_+_5410607 | 0.78 |
ENST00000328611.3
|
OR51M1
|
olfactory receptor, family 51, subfamily M, member 1 |
| chr13_-_110438914 | 0.78 |
ENST00000375856.3
|
IRS2
|
insulin receptor substrate 2 |
| chr11_-_101000445 | 0.78 |
ENST00000534013.1
|
PGR
|
progesterone receptor |
| chr10_-_101945771 | 0.77 |
ENST00000370408.2
ENST00000407654.3 |
ERLIN1
|
ER lipid raft associated 1 |
| chr3_+_124223586 | 0.76 |
ENST00000393496.1
|
KALRN
|
kalirin, RhoGEF kinase |
| chr21_+_17791648 | 0.76 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr9_-_5833027 | 0.75 |
ENST00000339450.5
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr12_-_10959892 | 0.75 |
ENST00000240615.2
|
TAS2R8
|
taste receptor, type 2, member 8 |
| chr8_-_19459993 | 0.75 |
ENST00000454498.2
ENST00000520003.1 |
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr4_-_138453606 | 0.75 |
ENST00000412923.2
ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18
|
protocadherin 18 |
| chr19_+_45844032 | 0.74 |
ENST00000589837.1
|
KLC3
|
kinesin light chain 3 |
| chr7_+_63361201 | 0.74 |
ENST00000450544.1
|
RP11-340I6.8
|
RP11-340I6.8 |
| chr3_+_51895621 | 0.74 |
ENST00000333127.3
|
IQCF2
|
IQ motif containing F2 |
| chr17_+_56769924 | 0.74 |
ENST00000461271.1
ENST00000583539.1 ENST00000337432.4 ENST00000421782.2 |
RAD51C
|
RAD51 paralog C |
| chr6_+_28249299 | 0.73 |
ENST00000405948.2
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr8_+_62747349 | 0.73 |
ENST00000517953.1
ENST00000520097.1 ENST00000519766.1 |
RP11-705O24.1
|
RP11-705O24.1 |
| chr6_+_28249332 | 0.72 |
ENST00000259883.3
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr21_+_17791838 | 0.72 |
ENST00000453910.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr3_-_165555200 | 0.69 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr2_+_179149636 | 0.69 |
ENST00000409631.1
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr4_-_48782259 | 0.68 |
ENST00000507711.1
ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL
|
FRY-like |
| chr4_-_110624564 | 0.68 |
ENST00000352981.3
ENST00000265164.2 ENST00000505486.1 |
CASP6
|
caspase 6, apoptosis-related cysteine peptidase |
| chr6_+_72922590 | 0.67 |
ENST00000523963.1
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr2_+_108994633 | 0.66 |
ENST00000409309.3
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr8_+_134125727 | 0.66 |
ENST00000521107.1
|
TG
|
thyroglobulin |
| chr2_-_68290106 | 0.65 |
ENST00000407324.1
ENST00000355848.3 ENST00000409302.1 ENST00000410067.3 |
C1D
|
C1D nuclear receptor corepressor |
| chr11_-_102576537 | 0.65 |
ENST00000260229.4
|
MMP27
|
matrix metallopeptidase 27 |
| chr6_+_73076432 | 0.64 |
ENST00000414192.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr10_-_95241951 | 0.64 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr3_+_16216137 | 0.63 |
ENST00000339732.5
|
GALNT15
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
| chr5_+_175288631 | 0.63 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr1_-_86861660 | 0.63 |
ENST00000486215.1
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr22_+_29702996 | 0.63 |
ENST00000406549.3
ENST00000360113.2 ENST00000341313.6 ENST00000403764.1 ENST00000471961.1 ENST00000407854.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chr6_+_116691001 | 0.62 |
ENST00000537543.1
|
DSE
|
dermatan sulfate epimerase |
| chr2_+_1417228 | 0.62 |
ENST00000382269.3
ENST00000337415.3 ENST00000345913.4 ENST00000346956.3 ENST00000349624.3 ENST00000539820.1 ENST00000329066.4 ENST00000382201.3 |
TPO
|
thyroid peroxidase |
| chr10_+_81462983 | 0.61 |
ENST00000448135.1
ENST00000429828.1 ENST00000372321.1 |
NUTM2B
|
NUT family member 2B |
| chr2_-_99279928 | 0.61 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr2_+_109237717 | 0.61 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr5_-_9630463 | 0.61 |
ENST00000382492.2
|
TAS2R1
|
taste receptor, type 2, member 1 |
| chr7_-_72992865 | 0.60 |
ENST00000452475.1
|
TBL2
|
transducin (beta)-like 2 |
| chrX_+_55744228 | 0.60 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
| chr1_-_21377383 | 0.59 |
ENST00000374935.3
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr11_+_101983176 | 0.59 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr12_-_10978957 | 0.59 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr7_+_150382781 | 0.59 |
ENST00000223293.5
ENST00000474605.1 |
GIMAP2
|
GTPase, IMAP family member 2 |
| chr12_-_25055177 | 0.58 |
ENST00000538118.1
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr14_-_74551096 | 0.58 |
ENST00000350259.4
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr11_-_124981475 | 0.58 |
ENST00000532156.1
ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr15_+_65843130 | 0.57 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr7_-_121784285 | 0.57 |
ENST00000417368.2
|
AASS
|
aminoadipate-semialdehyde synthase |
| chr6_+_148663729 | 0.56 |
ENST00000367467.3
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr14_+_31046959 | 0.56 |
ENST00000547532.1
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr6_+_72922505 | 0.56 |
ENST00000401910.3
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr7_+_138943265 | 0.55 |
ENST00000483726.1
|
UBN2
|
ubinuclein 2 |
| chr1_-_174992544 | 0.55 |
ENST00000476371.1
|
MRPS14
|
mitochondrial ribosomal protein S14 |
| chr2_-_89247338 | 0.55 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5 |
| chr17_+_66521936 | 0.55 |
ENST00000592800.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr16_-_66584059 | 0.55 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr21_-_31869451 | 0.54 |
ENST00000334058.2
|
KRTAP19-4
|
keratin associated protein 19-4 |
| chr7_+_80275953 | 0.54 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr12_+_101988627 | 0.54 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr5_-_93447333 | 0.53 |
ENST00000395965.3
ENST00000505869.1 ENST00000509163.1 |
FAM172A
|
family with sequence similarity 172, member A |
| chr6_+_32605195 | 0.53 |
ENST00000374949.2
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chrX_-_154033793 | 0.52 |
ENST00000369534.3
ENST00000413259.3 |
MPP1
|
membrane protein, palmitoylated 1, 55kDa |
| chr8_-_93978309 | 0.52 |
ENST00000517858.1
ENST00000378861.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr17_-_76870222 | 0.52 |
ENST00000585421.1
|
TIMP2
|
TIMP metallopeptidase inhibitor 2 |
| chr8_-_17752996 | 0.51 |
ENST00000381841.2
ENST00000427924.1 |
FGL1
|
fibrinogen-like 1 |
| chrX_+_55744166 | 0.51 |
ENST00000374941.4
ENST00000414239.1 |
RRAGB
|
Ras-related GTP binding B |
| chr2_+_28618532 | 0.51 |
ENST00000545753.1
|
FOSL2
|
FOS-like antigen 2 |
| chr10_+_101491968 | 0.51 |
ENST00000370476.5
ENST00000370472.4 |
CUTC
|
cutC copper transporter |
| chr9_+_21409146 | 0.51 |
ENST00000380205.1
|
IFNA8
|
interferon, alpha 8 |
| chr6_-_158589259 | 0.51 |
ENST00000367101.1
ENST00000367104.3 ENST00000367102.2 |
SERAC1
|
serine active site containing 1 |
| chr1_+_196788887 | 0.50 |
ENST00000320493.5
ENST00000367424.4 ENST00000367421.3 |
CFHR1
CFHR2
|
complement factor H-related 1 complement factor H-related 2 |
| chr17_-_76870126 | 0.50 |
ENST00000586057.1
|
TIMP2
|
TIMP metallopeptidase inhibitor 2 |
| chr5_-_42812143 | 0.50 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr16_-_90096309 | 0.49 |
ENST00000408886.2
|
C16orf3
|
chromosome 16 open reading frame 3 |
| chr1_-_45140074 | 0.49 |
ENST00000420706.1
ENST00000372235.3 ENST00000372242.3 ENST00000372243.3 ENST00000372244.3 |
TMEM53
|
transmembrane protein 53 |
| chr2_+_108994466 | 0.49 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr4_+_83956312 | 0.49 |
ENST00000509317.1
ENST00000503682.1 ENST00000511653.1 |
COPS4
|
COP9 signalosome subunit 4 |
| chr11_+_92085262 | 0.49 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr5_-_94417314 | 0.49 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr12_+_29376673 | 0.48 |
ENST00000547116.1
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr11_+_34643600 | 0.48 |
ENST00000530286.1
ENST00000533754.1 |
EHF
|
ets homologous factor |
| chr19_+_58193388 | 0.48 |
ENST00000596085.1
ENST00000594684.1 |
ZNF551
AC003006.7
|
zinc finger protein 551 Uncharacterized protein |
| chr7_+_107224364 | 0.47 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr2_+_24272543 | 0.47 |
ENST00000380991.4
|
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa |
| chr4_+_83956237 | 0.47 |
ENST00000264389.2
|
COPS4
|
COP9 signalosome subunit 4 |
| chr21_-_40033618 | 0.47 |
ENST00000417133.2
ENST00000398910.1 ENST00000442448.1 |
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chrX_+_36053908 | 0.47 |
ENST00000378660.2
|
CHDC2
|
calponin homology domain containing 2 |
| chr8_-_17752912 | 0.46 |
ENST00000398054.1
ENST00000381840.2 |
FGL1
|
fibrinogen-like 1 |
| chr3_-_105588231 | 0.46 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr15_+_58702742 | 0.46 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr14_-_23426322 | 0.46 |
ENST00000555367.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr8_-_93978333 | 0.46 |
ENST00000524037.1
ENST00000520430.1 ENST00000521617.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr8_-_93978346 | 0.46 |
ENST00000523580.1
|
TRIQK
|
triple QxxK/R motif containing |
| chr12_+_29376592 | 0.45 |
ENST00000182377.4
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr4_-_110723194 | 0.45 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr1_-_150978953 | 0.45 |
ENST00000493834.2
|
FAM63A
|
family with sequence similarity 63, member A |
| chr12_+_29302119 | 0.45 |
ENST00000536681.3
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr9_+_27109392 | 0.45 |
ENST00000406359.4
|
TEK
|
TEK tyrosine kinase, endothelial |
| chrX_-_57164058 | 0.45 |
ENST00000374906.3
|
SPIN2A
|
spindlin family, member 2A |
| chr1_+_43291220 | 0.44 |
ENST00000372514.3
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group) |
| chr17_-_21156578 | 0.44 |
ENST00000399011.2
ENST00000468196.1 |
C17orf103
|
chromosome 17 open reading frame 103 |
| chr6_+_27791862 | 0.44 |
ENST00000355057.1
|
HIST1H4J
|
histone cluster 1, H4j |
| chr4_-_110723134 | 0.44 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr10_+_13203543 | 0.43 |
ENST00000378714.3
ENST00000479669.1 ENST00000484800.2 |
MCM10
|
minichromosome maintenance complex component 10 |
| chrX_-_16730984 | 0.43 |
ENST00000380241.3
|
CTPS2
|
CTP synthase 2 |
| chr17_+_56315787 | 0.43 |
ENST00000262290.4
ENST00000421678.2 |
LPO
|
lactoperoxidase |
| chr14_+_96000930 | 0.43 |
ENST00000331334.4
|
GLRX5
|
glutaredoxin 5 |
| chr8_-_121457332 | 0.43 |
ENST00000518918.1
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr16_+_20775024 | 0.43 |
ENST00000289416.5
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr19_+_15852203 | 0.43 |
ENST00000305892.1
|
OR10H3
|
olfactory receptor, family 10, subfamily H, member 3 |
| chrX_-_16730688 | 0.42 |
ENST00000359276.4
|
CTPS2
|
CTP synthase 2 |
| chr7_+_101460882 | 0.42 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr10_-_91011548 | 0.42 |
ENST00000456827.1
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase |
| chr17_-_79105734 | 0.42 |
ENST00000417379.1
|
AATK
|
apoptosis-associated tyrosine kinase |
| chr6_+_32709119 | 0.42 |
ENST00000374940.3
|
HLA-DQA2
|
major histocompatibility complex, class II, DQ alpha 2 |
| chr12_+_52695617 | 0.42 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr12_+_125549925 | 0.41 |
ENST00000316519.6
|
AACS
|
acetoacetyl-CoA synthetase |
| chr1_+_367640 | 0.41 |
ENST00000426406.1
|
OR4F29
|
olfactory receptor, family 4, subfamily F, member 29 |
| chr3_-_114343039 | 0.41 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr12_+_58087901 | 0.41 |
ENST00000315970.7
ENST00000547079.1 ENST00000439210.2 ENST00000389146.6 ENST00000413095.2 ENST00000551035.1 ENST00000257966.8 ENST00000435406.2 ENST00000550372.1 ENST00000389142.5 |
OS9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr9_+_130911770 | 0.41 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr12_+_101988774 | 0.40 |
ENST00000545503.2
ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr2_+_90121477 | 0.40 |
ENST00000483379.1
|
IGKV1D-17
|
immunoglobulin kappa variable 1D-17 |
| chr14_+_23012122 | 0.40 |
ENST00000390534.1
|
TRAJ3
|
T cell receptor alpha joining 3 |
| chr20_-_29978286 | 0.40 |
ENST00000376315.2
|
DEFB119
|
defensin, beta 119 |
| chr6_-_35656685 | 0.40 |
ENST00000539068.1
ENST00000540787.1 |
FKBP5
|
FK506 binding protein 5 |
| chr19_+_10222189 | 0.40 |
ENST00000321826.4
|
P2RY11
|
purinergic receptor P2Y, G-protein coupled, 11 |
| chr11_-_31531121 | 0.40 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr19_+_13842559 | 0.40 |
ENST00000586600.1
|
CCDC130
|
coiled-coil domain containing 130 |
| chr16_+_33204156 | 0.40 |
ENST00000398667.4
|
TP53TG3C
|
TP53 target 3C |
| chr6_-_116575226 | 0.40 |
ENST00000420283.1
|
TSPYL4
|
TSPY-like 4 |
| chr19_+_1041212 | 0.39 |
ENST00000433129.1
|
ABCA7
|
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr11_+_57308979 | 0.39 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr14_+_22433675 | 0.39 |
ENST00000390442.3
|
TRAV12-3
|
T cell receptor alpha variable 12-3 |
| chr5_-_176433350 | 0.38 |
ENST00000377227.4
ENST00000377219.2 |
UIMC1
|
ubiquitin interaction motif containing 1 |
| chr1_-_35658736 | 0.38 |
ENST00000357214.5
|
SFPQ
|
splicing factor proline/glutamine-rich |
| chr10_-_18948156 | 0.38 |
ENST00000414939.1
ENST00000449529.1 ENST00000456217.1 ENST00000444660.1 |
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr5_+_140220769 | 0.38 |
ENST00000531613.1
ENST00000378123.3 |
PCDHA8
|
protocadherin alpha 8 |
| chrX_+_47444613 | 0.37 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr2_-_89292422 | 0.37 |
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8 |
| chr19_-_36001113 | 0.37 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr16_-_19897455 | 0.37 |
ENST00000568214.1
ENST00000569479.1 |
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B |
| chr15_-_55700457 | 0.36 |
ENST00000442196.3
ENST00000563171.1 ENST00000425574.3 |
CCPG1
|
cell cycle progression 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HMX3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.1 | GO:1903285 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.5 | 2.8 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.3 | 2.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.3 | 0.9 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.3 | 0.9 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.3 | 0.8 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.2 | 1.4 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.2 | 1.0 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.2 | 0.8 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.2 | 1.0 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.2 | 0.8 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 2.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 1.0 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.6 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 0.5 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.4 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.7 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.7 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.4 | GO:1902995 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.1 | 0.8 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 | 0.9 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.8 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 1.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.3 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 2.1 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 0.6 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 1.0 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 0.8 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.8 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 0.9 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.1 | 0.3 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.1 | 0.6 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 1.4 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.3 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.1 | 0.5 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 1.0 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 | 0.7 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.7 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 0.6 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 1.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.5 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.2 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.5 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.2 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.1 | 0.3 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 0.4 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.1 | 2.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.2 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.1 | 0.2 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.4 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.3 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.1 | 0.3 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.1 | 0.3 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.1 | 0.9 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.4 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 1.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.3 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 0.3 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 0.4 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 0.2 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.1 | 0.6 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 | 0.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.2 | GO:0050894 | determination of affect(GO:0050894) |
| 0.1 | 0.4 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 0.4 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.2 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.1 | 0.3 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.1 | 0.2 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 1.8 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.3 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.7 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.9 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.0 | 0.1 | GO:0048867 | ganglion mother cell fate determination(GO:0007402) stem cell fate determination(GO:0048867) |
| 0.0 | 0.2 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.3 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.5 | GO:0098870 | neuronal action potential propagation(GO:0019227) positive regulation of axon regeneration(GO:0048680) action potential propagation(GO:0098870) |
| 0.0 | 0.2 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.5 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.2 | GO:1904995 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.3 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.2 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.2 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.4 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.0 | 0.2 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.2 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.0 | 0.0 | GO:1902908 | regulation of melanosome transport(GO:1902908) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.9 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.0 | 0.2 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.3 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 0.4 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.1 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 1.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.3 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.3 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.6 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.0 | 0.6 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 1.7 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 1.0 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.2 | GO:0051552 | coumarin metabolic process(GO:0009804) flavone metabolic process(GO:0051552) |
| 0.0 | 0.2 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.1 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.6 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.5 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.7 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.6 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.5 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.6 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 3.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.1 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.2 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.1 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.0 | 0.4 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.1 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.3 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.0 | 0.3 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.5 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.5 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.6 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.2 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.1 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.1 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.5 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.1 | GO:1904430 | cellular hyperosmotic salinity response(GO:0071475) negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0035743 | CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.6 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.3 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.2 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.6 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.0 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.0 | 0.5 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 2.3 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.5 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.1 | GO:0001781 | neutrophil apoptotic process(GO:0001781) regulation of neutrophil apoptotic process(GO:0033029) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 1.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 0.7 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.6 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 1.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 2.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.7 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.6 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 1.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.5 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 0.7 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 1.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.4 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 1.0 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 0.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 1.0 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 1.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.7 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.4 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.4 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 1.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 1.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.1 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.4 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.5 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 2.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.0 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 2.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 1.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 2.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.3 | 1.0 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.3 | 0.9 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.3 | 0.9 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.3 | 1.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.3 | 0.8 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.2 | 0.5 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.2 | 0.7 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.2 | 1.9 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.2 | 1.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.4 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.6 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.8 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 2.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.6 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.1 | 0.3 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.6 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.3 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.1 | 2.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.5 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 1.0 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 1.0 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.3 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.3 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.1 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 1.0 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.7 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.2 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.3 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.3 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 0.3 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 0.2 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.1 | 0.3 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.1 | 0.2 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 0.3 | GO:0070404 | NADH binding(GO:0070404) |
| 0.1 | 0.2 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.2 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.1 | 0.2 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.1 | 0.2 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 1.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 0.2 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.1 | 0.2 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.1 | 0.8 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 1.2 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 1.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.4 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.0 | 1.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 1.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.6 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 2.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.3 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.3 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.0 | 0.1 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.6 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.7 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 1.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.0 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.9 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 2.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.0 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 7.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.8 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.9 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 2.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 2.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 5.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 1.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.8 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 1.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.0 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 1.1 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.6 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.1 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.5 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 1.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.8 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |