Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
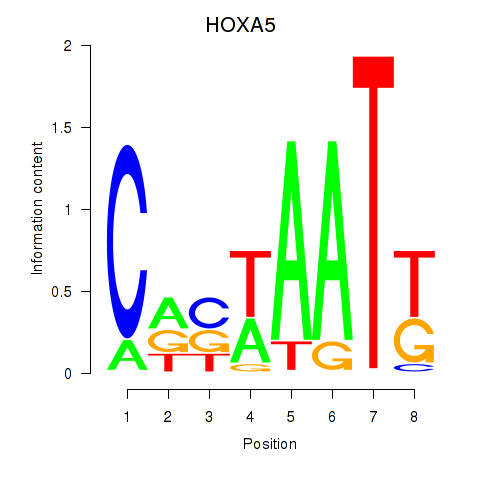
Results for HOXA5
Z-value: 0.52
Transcription factors associated with HOXA5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA5
|
ENSG00000106004.4 | homeobox A5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA5 | hg19_v2_chr7_-_27183263_27183287 | 0.12 | 5.6e-01 | Click! |
Activity profile of HOXA5 motif
Sorted Z-values of HOXA5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_100111580 | 1.95 |
ENST00000605497.1
|
PALMD
|
palmdelphin |
| chr1_+_100111479 | 1.89 |
ENST00000263174.4
|
PALMD
|
palmdelphin |
| chrX_+_54947229 | 1.57 |
ENST00000442098.1
ENST00000430420.1 ENST00000453081.1 ENST00000173898.7 ENST00000319167.8 ENST00000375022.4 ENST00000399736.1 ENST00000440072.1 ENST00000420798.2 ENST00000431115.1 ENST00000440759.1 ENST00000375041.2 |
TRO
|
trophinin |
| chr14_-_92413353 | 1.46 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr1_+_84609944 | 1.08 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr14_-_92413727 | 0.96 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr1_+_221051699 | 0.94 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr20_+_4667094 | 0.64 |
ENST00000424424.1
ENST00000457586.1 |
PRNP
|
prion protein |
| chr6_+_155537771 | 0.55 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr7_+_99717230 | 0.53 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr14_+_65878565 | 0.52 |
ENST00000556518.1
ENST00000557164.1 |
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr1_-_227505289 | 0.47 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr9_+_75766763 | 0.44 |
ENST00000456643.1
ENST00000415424.1 |
ANXA1
|
annexin A1 |
| chr19_-_1021113 | 0.43 |
ENST00000333175.5
ENST00000356663.3 |
TMEM259
|
transmembrane protein 259 |
| chr17_-_27224621 | 0.43 |
ENST00000394906.2
ENST00000585169.1 ENST00000394908.4 |
FLOT2
|
flotillin 2 |
| chr11_-_31531121 | 0.41 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr6_+_130339710 | 0.41 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chrX_+_10124977 | 0.37 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr17_-_295730 | 0.35 |
ENST00000329099.4
|
FAM101B
|
family with sequence similarity 101, member B |
| chr21_-_40033618 | 0.34 |
ENST00000417133.2
ENST00000398910.1 ENST00000442448.1 |
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chrX_+_55478538 | 0.32 |
ENST00000342972.1
|
MAGEH1
|
melanoma antigen family H, 1 |
| chr6_-_143266297 | 0.30 |
ENST00000367603.2
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr14_+_21156915 | 0.28 |
ENST00000397990.4
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr20_+_4666882 | 0.28 |
ENST00000379440.4
ENST00000430350.2 |
PRNP
|
prion protein |
| chr3_+_127770455 | 0.27 |
ENST00000464451.1
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr11_-_2162468 | 0.27 |
ENST00000434045.2
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr19_+_53761545 | 0.27 |
ENST00000341702.3
|
VN1R2
|
vomeronasal 1 receptor 2 |
| chr12_+_74931551 | 0.27 |
ENST00000519948.2
|
ATXN7L3B
|
ataxin 7-like 3B |
| chr19_+_50016610 | 0.27 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr14_+_102276192 | 0.26 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr6_-_116575226 | 0.26 |
ENST00000420283.1
|
TSPYL4
|
TSPY-like 4 |
| chr8_-_141774467 | 0.26 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr1_-_244006528 | 0.24 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr6_-_154677900 | 0.23 |
ENST00000265198.4
ENST00000520261.1 |
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr3_-_143567262 | 0.23 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr3_-_149093499 | 0.23 |
ENST00000472441.1
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chrX_-_118826784 | 0.23 |
ENST00000394616.4
|
SEPT6
|
septin 6 |
| chr7_-_99717463 | 0.21 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr2_-_89292422 | 0.21 |
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8 |
| chr5_+_140552218 | 0.21 |
ENST00000231137.3
|
PCDHB7
|
protocadherin beta 7 |
| chr10_+_103986085 | 0.20 |
ENST00000370005.3
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chr2_-_61697862 | 0.20 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr22_+_46476192 | 0.20 |
ENST00000443490.1
|
FLJ27365
|
hsa-mir-4763 |
| chr2_+_114163945 | 0.20 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr6_+_42896865 | 0.18 |
ENST00000372836.4
ENST00000394142.3 |
CNPY3
|
canopy FGF signaling regulator 3 |
| chr16_-_18573396 | 0.18 |
ENST00000543392.1
ENST00000381474.3 ENST00000330537.6 |
NOMO2
|
NODAL modulator 2 |
| chr19_+_8455077 | 0.18 |
ENST00000328024.6
|
RAB11B
|
RAB11B, member RAS oncogene family |
| chr1_+_153950202 | 0.17 |
ENST00000608236.1
|
RP11-422P24.11
|
RP11-422P24.11 |
| chr2_-_158300556 | 0.17 |
ENST00000264192.3
|
CYTIP
|
cytohesin 1 interacting protein |
| chr6_-_32157947 | 0.16 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr17_+_74733744 | 0.16 |
ENST00000586689.1
ENST00000587661.1 ENST00000593181.1 ENST00000336509.4 ENST00000355954.3 |
MFSD11
|
major facilitator superfamily domain containing 11 |
| chr17_+_74734052 | 0.16 |
ENST00000590514.1
|
MFSD11
|
major facilitator superfamily domain containing 11 |
| chr14_+_67291158 | 0.16 |
ENST00000555456.1
|
GPHN
|
gephyrin |
| chr3_+_44607051 | 0.16 |
ENST00000419137.1
|
RP11-944L7.5
|
Zinc finger protein with KRAB and SCAN domains 7 |
| chr4_+_74275057 | 0.15 |
ENST00000511370.1
|
ALB
|
albumin |
| chr16_+_14927538 | 0.15 |
ENST00000287667.7
|
NOMO1
|
NODAL modulator 1 |
| chr19_-_51522955 | 0.14 |
ENST00000358789.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr1_+_152943122 | 0.14 |
ENST00000328051.2
|
SPRR4
|
small proline-rich protein 4 |
| chr6_-_2751146 | 0.14 |
ENST00000268446.5
ENST00000274643.7 |
MYLK4
|
myosin light chain kinase family, member 4 |
| chr8_+_75262629 | 0.14 |
ENST00000434412.2
|
GDAP1
|
ganglioside induced differentiation associated protein 1 |
| chr2_-_179343226 | 0.14 |
ENST00000434643.2
|
FKBP7
|
FK506 binding protein 7 |
| chr13_+_97874574 | 0.14 |
ENST00000343600.4
ENST00000345429.6 ENST00000376673.3 |
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr6_+_140175987 | 0.14 |
ENST00000414038.1
ENST00000431609.1 |
RP5-899B16.1
|
RP5-899B16.1 |
| chr11_-_33744487 | 0.14 |
ENST00000426650.2
|
CD59
|
CD59 molecule, complement regulatory protein |
| chr15_-_34628951 | 0.13 |
ENST00000397707.2
ENST00000560611.1 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr5_+_140723601 | 0.13 |
ENST00000253812.6
|
PCDHGA3
|
protocadherin gamma subfamily A, 3 |
| chr12_+_104337515 | 0.12 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr11_-_46722117 | 0.12 |
ENST00000311956.4
|
ARHGAP1
|
Rho GTPase activating protein 1 |
| chr10_-_115904361 | 0.12 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr12_-_91574142 | 0.12 |
ENST00000547937.1
|
DCN
|
decorin |
| chr3_+_150126101 | 0.11 |
ENST00000361875.3
ENST00000361136.2 |
TSC22D2
|
TSC22 domain family, member 2 |
| chr12_+_25205446 | 0.11 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr7_-_99716952 | 0.10 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr2_+_90259593 | 0.10 |
ENST00000471857.1
|
IGKV1D-8
|
immunoglobulin kappa variable 1D-8 |
| chr13_+_76378305 | 0.09 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chr15_-_73925651 | 0.09 |
ENST00000545878.1
ENST00000287226.8 ENST00000345330.4 |
NPTN
|
neuroplastin |
| chr6_+_167525277 | 0.09 |
ENST00000400926.2
|
CCR6
|
chemokine (C-C motif) receptor 6 |
| chr3_+_179322481 | 0.09 |
ENST00000259037.3
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr7_-_92855762 | 0.08 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr2_+_166095898 | 0.08 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chrX_+_99899180 | 0.08 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr20_-_34638841 | 0.08 |
ENST00000565493.1
|
LINC00657
|
long intergenic non-protein coding RNA 657 |
| chr5_+_69321074 | 0.08 |
ENST00000380751.5
ENST00000380750.3 ENST00000503931.1 ENST00000506542.1 |
SERF1B
|
small EDRK-rich factor 1B (centromeric) |
| chr5_-_55412774 | 0.07 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr14_+_102276132 | 0.07 |
ENST00000350249.3
ENST00000557621.1 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr11_-_33744256 | 0.07 |
ENST00000415002.2
ENST00000437761.2 ENST00000445143.2 |
CD59
|
CD59 molecule, complement regulatory protein |
| chr7_+_155437341 | 0.07 |
ENST00000401878.3
ENST00000392759.3 |
RBM33
|
RNA binding motif protein 33 |
| chrX_-_65253506 | 0.07 |
ENST00000427538.1
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr11_+_9685604 | 0.07 |
ENST00000447399.2
ENST00000318950.6 |
SWAP70
|
SWAP switching B-cell complex 70kDa subunit |
| chrX_+_75648046 | 0.07 |
ENST00000361470.2
|
MAGEE1
|
melanoma antigen family E, 1 |
| chr2_-_160919112 | 0.07 |
ENST00000283243.7
ENST00000392771.1 |
PLA2R1
|
phospholipase A2 receptor 1, 180kDa |
| chr4_-_152682129 | 0.07 |
ENST00000512306.1
ENST00000508611.1 ENST00000515812.1 ENST00000263985.6 |
PET112
|
PET112 homolog (yeast) |
| chr6_+_125304502 | 0.06 |
ENST00000519799.1
ENST00000368414.2 ENST00000359704.2 |
RNF217
|
ring finger protein 217 |
| chr1_+_177140633 | 0.06 |
ENST00000361539.4
|
BRINP2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr6_+_29426230 | 0.06 |
ENST00000442615.1
|
OR2H1
|
olfactory receptor, family 2, subfamily H, member 1 |
| chr8_+_92261516 | 0.06 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr8_-_135522425 | 0.06 |
ENST00000521673.1
|
ZFAT
|
zinc finger and AT hook domain containing |
| chr8_+_55528627 | 0.06 |
ENST00000220676.1
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant) |
| chr5_+_66300446 | 0.06 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr7_+_100136811 | 0.06 |
ENST00000300176.4
ENST00000262935.4 |
AGFG2
|
ArfGAP with FG repeats 2 |
| chr4_+_1283639 | 0.06 |
ENST00000303400.4
ENST00000505177.2 ENST00000503653.1 ENST00000264750.6 ENST00000502558.1 ENST00000452175.2 ENST00000514708.1 |
MAEA
|
macrophage erythroblast attacher |
| chr1_-_154150651 | 0.06 |
ENST00000302206.5
|
TPM3
|
tropomyosin 3 |
| chr15_-_67351586 | 0.06 |
ENST00000558071.1
|
RP11-798K3.2
|
RP11-798K3.2 |
| chr3_+_37035289 | 0.06 |
ENST00000455445.2
ENST00000441265.1 ENST00000435176.1 ENST00000429117.1 ENST00000536378.1 |
MLH1
|
mutL homolog 1 |
| chr1_+_150337100 | 0.06 |
ENST00000401000.4
|
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr18_+_13611431 | 0.05 |
ENST00000587757.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr10_+_17270214 | 0.05 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr9_-_110251836 | 0.05 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr16_+_10971037 | 0.05 |
ENST00000324288.8
ENST00000381835.5 |
CIITA
|
class II, major histocompatibility complex, transactivator |
| chr10_+_126630692 | 0.05 |
ENST00000359653.4
|
ZRANB1
|
zinc finger, RAN-binding domain containing 1 |
| chr1_+_150337144 | 0.05 |
ENST00000539519.1
ENST00000369067.3 ENST00000369068.4 |
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr1_-_63988846 | 0.05 |
ENST00000283568.8
ENST00000371092.3 ENST00000271002.10 |
ITGB3BP
|
integrin beta 3 binding protein (beta3-endonexin) |
| chr4_+_71063641 | 0.05 |
ENST00000514097.1
|
ODAM
|
odontogenic, ameloblast asssociated |
| chr4_-_103749205 | 0.05 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr9_+_131037623 | 0.04 |
ENST00000495313.1
ENST00000372898.2 |
SWI5
|
SWI5 recombination repair homolog (yeast) |
| chr6_+_124125286 | 0.04 |
ENST00000368416.1
ENST00000368417.1 ENST00000546092.1 |
NKAIN2
|
Na+/K+ transporting ATPase interacting 2 |
| chr1_+_154229547 | 0.04 |
ENST00000428595.1
|
UBAP2L
|
ubiquitin associated protein 2-like |
| chr14_+_102276209 | 0.04 |
ENST00000445439.3
ENST00000334743.5 ENST00000557095.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr3_-_157221128 | 0.04 |
ENST00000392833.2
ENST00000362010.2 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr2_+_74154032 | 0.03 |
ENST00000356837.6
|
DGUOK
|
deoxyguanosine kinase |
| chr10_-_75226166 | 0.03 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr3_+_174577070 | 0.03 |
ENST00000454872.1
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2 |
| chr4_+_158493642 | 0.03 |
ENST00000507108.1
ENST00000455598.1 ENST00000509450.1 |
RP11-364P22.1
|
RP11-364P22.1 |
| chr13_+_76378407 | 0.03 |
ENST00000447038.1
|
LMO7
|
LIM domain 7 |
| chr8_-_90769422 | 0.03 |
ENST00000524190.1
ENST00000523859.1 |
RP11-37B2.1
|
RP11-37B2.1 |
| chr7_+_26438187 | 0.03 |
ENST00000439120.1
ENST00000430548.1 ENST00000421862.1 ENST00000449537.1 ENST00000420774.1 ENST00000418758.2 |
AC004540.5
|
AC004540.5 |
| chr12_-_54653313 | 0.03 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chrX_+_102024075 | 0.03 |
ENST00000431616.1
ENST00000440496.1 ENST00000420471.1 ENST00000435966.1 |
LINC00630
|
long intergenic non-protein coding RNA 630 |
| chr4_-_168155169 | 0.03 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr10_-_97200772 | 0.02 |
ENST00000371241.1
ENST00000354106.3 ENST00000371239.1 ENST00000361941.3 ENST00000277982.5 ENST00000371245.3 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chrX_+_72062802 | 0.02 |
ENST00000373533.1
|
DMRTC1B
|
DMRT-like family C1B |
| chr9_+_140125385 | 0.02 |
ENST00000361134.2
|
SLC34A3
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 3 |
| chr20_+_2361545 | 0.02 |
ENST00000202625.2
ENST00000381423.1 |
TGM6
|
transglutaminase 6 |
| chr3_+_196466710 | 0.02 |
ENST00000327134.3
|
PAK2
|
p21 protein (Cdc42/Rac)-activated kinase 2 |
| chrX_-_72097698 | 0.02 |
ENST00000373530.1
|
DMRTC1
|
DMRT-like family C1 |
| chr13_+_30510003 | 0.02 |
ENST00000400540.1
|
LINC00544
|
long intergenic non-protein coding RNA 544 |
| chr5_+_140207536 | 0.02 |
ENST00000529310.1
ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chr4_-_164534657 | 0.01 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr4_-_103746683 | 0.01 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr8_-_42234745 | 0.01 |
ENST00000220812.2
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr9_+_33290491 | 0.01 |
ENST00000379540.3
ENST00000379521.4 ENST00000318524.6 |
NFX1
|
nuclear transcription factor, X-box binding 1 |
| chr1_-_91487013 | 0.01 |
ENST00000347275.5
ENST00000370440.1 |
ZNF644
|
zinc finger protein 644 |
| chr19_-_35264089 | 0.00 |
ENST00000588760.1
ENST00000329285.8 ENST00000587354.2 |
ZNF599
|
zinc finger protein 599 |
| chr4_-_103746924 | 0.00 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:1990535 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) neuron projection maintenance(GO:1990535) |
| 0.2 | 2.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 0.5 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.2 | 1.4 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.2 | 1.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.3 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 0.4 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.1 | 0.3 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.2 | GO:0042040 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 | 0.2 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.1 | 0.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.3 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 1.5 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0051257 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.2 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.1 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.0 | 0.4 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 3.8 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.4 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.2 | GO:0005940 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.1 | 0.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 1.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 1.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.4 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 3.2 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.0 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.2 | 0.9 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.1 | 0.3 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.4 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 1.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.2 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 2.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |