Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for HOXA9
Z-value: 0.74
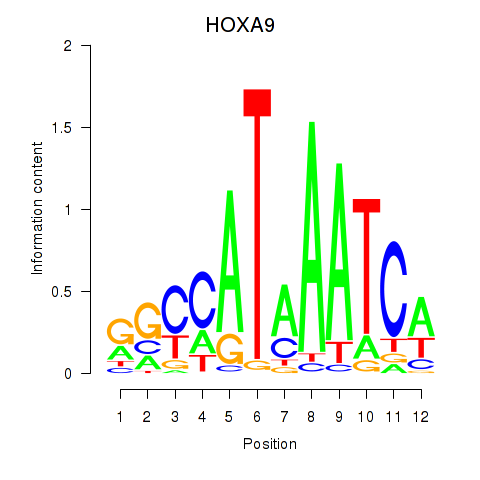
Transcription factors associated with HOXA9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA9
|
ENSG00000078399.11 | homeobox A9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA9 | hg19_v2_chr7_-_27205136_27205164 | -0.55 | 4.1e-03 | Click! |
Activity profile of HOXA9 motif
Sorted Z-values of HOXA9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_101185290 | 4.64 |
ENST00000370119.4
ENST00000347652.2 ENST00000294728.2 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr15_+_67418047 | 3.18 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr2_-_225811747 | 3.09 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr4_-_76944621 | 2.98 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr2_+_143635067 | 2.42 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr7_+_55177416 | 1.83 |
ENST00000450046.1
ENST00000454757.2 |
EGFR
|
epidermal growth factor receptor |
| chr10_+_91092241 | 1.72 |
ENST00000371811.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr4_-_69536346 | 1.62 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr8_+_31497271 | 1.54 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr6_+_126240442 | 1.50 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr1_-_173176452 | 1.49 |
ENST00000281834.3
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr2_+_143635222 | 1.49 |
ENST00000375773.2
ENST00000409512.1 ENST00000410015.2 |
KYNU
|
kynureninase |
| chr10_-_24770632 | 1.38 |
ENST00000596413.1
|
AL353583.1
|
AL353583.1 |
| chr21_+_37507210 | 1.28 |
ENST00000290354.5
|
CBR3
|
carbonyl reductase 3 |
| chr7_-_83824169 | 1.27 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr5_-_146833222 | 1.25 |
ENST00000534907.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr2_-_197226875 | 1.24 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr12_-_57472522 | 1.19 |
ENST00000379391.3
ENST00000300128.4 |
TMEM194A
|
transmembrane protein 194A |
| chr2_+_161993465 | 1.11 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chrX_+_9431324 | 1.02 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr8_-_20040638 | 0.91 |
ENST00000519026.1
ENST00000276373.5 ENST00000440926.1 ENST00000437980.1 |
SLC18A1
|
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr11_-_108464465 | 0.89 |
ENST00000525344.1
|
EXPH5
|
exophilin 5 |
| chr11_-_57092381 | 0.86 |
ENST00000358252.3
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr1_-_89591749 | 0.84 |
ENST00000370466.3
|
GBP2
|
guanylate binding protein 2, interferon-inducible |
| chr10_+_24755416 | 0.84 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr2_+_161993412 | 0.82 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr10_-_14050522 | 0.80 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr3_-_58613323 | 0.80 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr6_-_137539651 | 0.78 |
ENST00000543628.1
|
IFNGR1
|
interferon gamma receptor 1 |
| chr5_-_146833485 | 0.78 |
ENST00000398514.3
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr2_+_232063260 | 0.75 |
ENST00000349938.4
|
ARMC9
|
armadillo repeat containing 9 |
| chr4_+_39046615 | 0.75 |
ENST00000261425.3
ENST00000508137.2 |
KLHL5
|
kelch-like family member 5 |
| chr11_-_104905840 | 0.74 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr22_+_30477000 | 0.73 |
ENST00000403975.1
|
HORMAD2
|
HORMA domain containing 2 |
| chr6_+_18155560 | 0.72 |
ENST00000546309.2
ENST00000388870.2 ENST00000397244.1 |
KDM1B
|
lysine (K)-specific demethylase 1B |
| chr4_-_70080449 | 0.70 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr5_-_94417314 | 0.69 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr3_-_190167571 | 0.69 |
ENST00000354905.2
|
TMEM207
|
transmembrane protein 207 |
| chr8_-_10512569 | 0.68 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr15_+_63414760 | 0.67 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr11_-_126870655 | 0.66 |
ENST00000525144.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr4_+_146539415 | 0.65 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr2_+_232063436 | 0.65 |
ENST00000440107.1
|
ARMC9
|
armadillo repeat containing 9 |
| chr5_+_176731572 | 0.64 |
ENST00000503853.1
|
PRELID1
|
PRELI domain containing 1 |
| chr9_-_95640218 | 0.62 |
ENST00000395506.3
ENST00000375495.3 ENST00000332591.6 |
ZNF484
|
zinc finger protein 484 |
| chr4_-_48018580 | 0.61 |
ENST00000514170.1
|
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr8_-_20040601 | 0.61 |
ENST00000265808.7
ENST00000522513.1 |
SLC18A1
|
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr16_+_56691606 | 0.60 |
ENST00000334350.6
|
MT1F
|
metallothionein 1F |
| chrX_+_95939638 | 0.60 |
ENST00000373061.3
ENST00000373054.4 ENST00000355827.4 |
DIAPH2
|
diaphanous-related formin 2 |
| chrX_-_45629661 | 0.59 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr16_+_56691838 | 0.58 |
ENST00000394501.2
|
MT1F
|
metallothionein 1F |
| chr16_+_56691911 | 0.58 |
ENST00000568475.1
|
MT1F
|
metallothionein 1F |
| chrX_+_144908928 | 0.57 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr11_+_6866883 | 0.57 |
ENST00000299454.4
ENST00000379831.2 |
OR10A5
|
olfactory receptor, family 10, subfamily A, member 5 |
| chr1_+_50575292 | 0.57 |
ENST00000371821.1
ENST00000371819.1 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr1_+_50574585 | 0.55 |
ENST00000371824.1
ENST00000371823.4 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr6_+_26365443 | 0.55 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr14_+_22977587 | 0.54 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chr2_-_209028300 | 0.54 |
ENST00000304502.4
|
CRYGA
|
crystallin, gamma A |
| chr10_-_90712520 | 0.53 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr2_-_69098566 | 0.50 |
ENST00000295379.1
|
BMP10
|
bone morphogenetic protein 10 |
| chr22_-_30642782 | 0.50 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr17_-_37934466 | 0.49 |
ENST00000583368.1
|
IKZF3
|
IKAROS family zinc finger 3 (Aiolos) |
| chr12_+_21207503 | 0.48 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chrX_-_63615297 | 0.47 |
ENST00000374852.3
ENST00000453546.1 |
MTMR8
|
myotubularin related protein 8 |
| chr20_+_43211149 | 0.47 |
ENST00000372886.1
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr16_+_70613770 | 0.47 |
ENST00000429149.2
ENST00000563721.2 |
IL34
|
interleukin 34 |
| chr17_-_39165366 | 0.47 |
ENST00000391588.1
|
KRTAP3-1
|
keratin associated protein 3-1 |
| chr3_+_119298523 | 0.46 |
ENST00000357003.3
|
ADPRH
|
ADP-ribosylarginine hydrolase |
| chr2_+_108994466 | 0.46 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr3_-_112693865 | 0.46 |
ENST00000471858.1
ENST00000295863.4 ENST00000308611.3 |
CD200R1
|
CD200 receptor 1 |
| chr21_-_19775973 | 0.46 |
ENST00000284885.3
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr2_+_171571827 | 0.46 |
ENST00000375281.3
|
SP5
|
Sp5 transcription factor |
| chr21_-_30257669 | 0.45 |
ENST00000303775.5
ENST00000351429.3 |
N6AMT1
|
N-6 adenine-specific DNA methyltransferase 1 (putative) |
| chr5_+_68485433 | 0.44 |
ENST00000502689.1
|
CENPH
|
centromere protein H |
| chr7_-_27169801 | 0.44 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chrX_-_110513703 | 0.44 |
ENST00000324068.1
|
CAPN6
|
calpain 6 |
| chr11_-_5345582 | 0.43 |
ENST00000328813.2
|
OR51B2
|
olfactory receptor, family 51, subfamily B, member 2 |
| chr6_+_160327974 | 0.42 |
ENST00000252660.4
|
MAS1
|
MAS1 oncogene |
| chr2_-_190044480 | 0.41 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr16_+_67063855 | 0.40 |
ENST00000563939.2
|
CBFB
|
core-binding factor, beta subunit |
| chr11_+_5474638 | 0.39 |
ENST00000341449.2
|
OR51I2
|
olfactory receptor, family 51, subfamily I, member 2 |
| chr17_+_73663402 | 0.39 |
ENST00000355423.3
|
SAP30BP
|
SAP30 binding protein |
| chr12_-_121477039 | 0.39 |
ENST00000257570.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr9_-_134955246 | 0.38 |
ENST00000357028.2
ENST00000474263.1 ENST00000292035.5 |
MED27
|
mediator complex subunit 27 |
| chr2_-_130031335 | 0.37 |
ENST00000375987.3
|
AC079586.1
|
AC079586.1 |
| chr6_-_138539627 | 0.37 |
ENST00000527246.2
|
PBOV1
|
prostate and breast cancer overexpressed 1 |
| chr18_+_9102592 | 0.37 |
ENST00000318388.6
|
NDUFV2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| chr1_-_7913089 | 0.37 |
ENST00000361696.5
|
UTS2
|
urotensin 2 |
| chr11_-_22647350 | 0.37 |
ENST00000327470.3
|
FANCF
|
Fanconi anemia, complementation group F |
| chr2_+_108994633 | 0.37 |
ENST00000409309.3
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr9_-_95244781 | 0.36 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr10_+_15001430 | 0.36 |
ENST00000407572.1
|
MEIG1
|
meiosis/spermiogenesis associated 1 |
| chr2_+_27255806 | 0.36 |
ENST00000238788.9
ENST00000404032.3 |
TMEM214
|
transmembrane protein 214 |
| chr2_-_88427568 | 0.35 |
ENST00000393750.3
ENST00000295834.3 |
FABP1
|
fatty acid binding protein 1, liver |
| chr12_-_13248598 | 0.34 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr1_-_197169672 | 0.34 |
ENST00000367405.4
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chr3_-_195310802 | 0.34 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr6_+_108977520 | 0.34 |
ENST00000540898.1
|
FOXO3
|
forkhead box O3 |
| chr10_+_102222798 | 0.34 |
ENST00000343737.5
|
WNT8B
|
wingless-type MMTV integration site family, member 8B |
| chr16_+_3074002 | 0.33 |
ENST00000326266.8
ENST00000574549.1 ENST00000575576.1 ENST00000253952.9 |
THOC6
|
THO complex 6 homolog (Drosophila) |
| chr3_+_185300391 | 0.33 |
ENST00000545472.1
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr19_-_42133420 | 0.33 |
ENST00000221954.2
ENST00000600925.1 |
CEACAM4
|
carcinoembryonic antigen-related cell adhesion molecule 4 |
| chr7_+_128312346 | 0.32 |
ENST00000480462.1
ENST00000378704.3 ENST00000477515.1 |
FAM71F2
|
family with sequence similarity 71, member F2 |
| chr7_+_20370746 | 0.31 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr5_+_34915444 | 0.31 |
ENST00000336767.5
|
BRIX1
|
BRX1, biogenesis of ribosomes, homolog (S. cerevisiae) |
| chr17_-_7232585 | 0.31 |
ENST00000571887.1
ENST00000315614.7 ENST00000399464.2 ENST00000570460.1 |
NEURL4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr6_+_111195973 | 0.30 |
ENST00000368885.3
ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr12_+_122181529 | 0.30 |
ENST00000541467.1
|
TMEM120B
|
transmembrane protein 120B |
| chr15_+_59730348 | 0.30 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chr7_+_120628731 | 0.30 |
ENST00000310396.5
|
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chrX_+_95939711 | 0.29 |
ENST00000373049.4
ENST00000324765.8 |
DIAPH2
|
diaphanous-related formin 2 |
| chr1_-_85100703 | 0.28 |
ENST00000370624.1
|
C1orf180
|
chromosome 1 open reading frame 180 |
| chr18_+_45778672 | 0.28 |
ENST00000600091.1
|
AC091150.1
|
HCG1818186; Uncharacterized protein |
| chr7_+_119913688 | 0.28 |
ENST00000331113.4
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr2_-_209051727 | 0.28 |
ENST00000453017.1
ENST00000423952.2 |
C2orf80
|
chromosome 2 open reading frame 80 |
| chr4_-_74964904 | 0.28 |
ENST00000508487.2
|
CXCL2
|
chemokine (C-X-C motif) ligand 2 |
| chr2_+_27799389 | 0.28 |
ENST00000408964.2
|
C2orf16
|
chromosome 2 open reading frame 16 |
| chr3_-_108836977 | 0.27 |
ENST00000232603.5
|
MORC1
|
MORC family CW-type zinc finger 1 |
| chr1_+_44584522 | 0.27 |
ENST00000372299.3
|
KLF17
|
Kruppel-like factor 17 |
| chr4_-_159644507 | 0.27 |
ENST00000307720.3
|
PPID
|
peptidylprolyl isomerase D |
| chr2_+_89986318 | 0.27 |
ENST00000491977.1
|
IGKV2D-29
|
immunoglobulin kappa variable 2D-29 |
| chr18_-_72264805 | 0.27 |
ENST00000577806.1
|
LINC00909
|
long intergenic non-protein coding RNA 909 |
| chr3_+_108321623 | 0.27 |
ENST00000497905.1
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr4_+_113970772 | 0.26 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr17_+_60447579 | 0.26 |
ENST00000450662.2
|
EFCAB3
|
EF-hand calcium binding domain 3 |
| chr15_+_40643227 | 0.26 |
ENST00000448599.2
|
PHGR1
|
proline/histidine/glycine-rich 1 |
| chr6_-_9939552 | 0.26 |
ENST00000460363.2
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr12_+_70219052 | 0.26 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chr19_-_10420459 | 0.25 |
ENST00000403352.1
ENST00000403903.3 |
ZGLP1
|
zinc finger, GATA-like protein 1 |
| chr12_-_13248705 | 0.25 |
ENST00000396310.2
|
GSG1
|
germ cell associated 1 |
| chrX_-_13338518 | 0.25 |
ENST00000380622.2
|
ATXN3L
|
ataxin 3-like |
| chr1_+_117963209 | 0.25 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr1_-_36948879 | 0.25 |
ENST00000373106.1
ENST00000373104.1 ENST00000373103.1 |
CSF3R
|
colony stimulating factor 3 receptor (granulocyte) |
| chr12_-_13248732 | 0.25 |
ENST00000396302.3
|
GSG1
|
germ cell associated 1 |
| chr1_-_153085984 | 0.25 |
ENST00000468739.1
|
SPRR2F
|
small proline-rich protein 2F |
| chr6_-_25832254 | 0.25 |
ENST00000476801.1
ENST00000244527.4 ENST00000427328.1 |
SLC17A1
|
solute carrier family 17 (organic anion transporter), member 1 |
| chr5_-_101834617 | 0.25 |
ENST00000513675.1
ENST00000379807.3 |
SLCO6A1
|
solute carrier organic anion transporter family, member 6A1 |
| chr6_+_99282570 | 0.24 |
ENST00000328345.5
|
POU3F2
|
POU class 3 homeobox 2 |
| chr2_+_208423840 | 0.24 |
ENST00000539789.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr5_+_38845960 | 0.24 |
ENST00000502536.1
|
OSMR
|
oncostatin M receptor |
| chr8_+_24151620 | 0.24 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr19_+_50529212 | 0.24 |
ENST00000270617.3
ENST00000445728.3 ENST00000601364.1 |
ZNF473
|
zinc finger protein 473 |
| chr17_+_4853442 | 0.24 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr15_-_68497657 | 0.24 |
ENST00000448060.2
ENST00000467889.1 |
CALML4
|
calmodulin-like 4 |
| chr15_+_65134088 | 0.24 |
ENST00000323544.4
ENST00000437723.1 |
PLEKHO2
AC069368.3
|
pleckstrin homology domain containing, family O member 2 Uncharacterized protein |
| chr2_+_149894968 | 0.24 |
ENST00000409642.3
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr8_+_16884740 | 0.23 |
ENST00000318063.5
|
MICU3
|
mitochondrial calcium uptake family, member 3 |
| chr18_-_72265035 | 0.23 |
ENST00000585279.1
ENST00000580048.1 |
LINC00909
|
long intergenic non-protein coding RNA 909 |
| chr4_-_109090106 | 0.23 |
ENST00000379951.2
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr3_+_35721106 | 0.23 |
ENST00000474696.1
ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr5_-_101834712 | 0.23 |
ENST00000506729.1
ENST00000389019.3 ENST00000379810.1 |
SLCO6A1
|
solute carrier organic anion transporter family, member 6A1 |
| chr8_+_24151553 | 0.23 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr11_-_3400330 | 0.23 |
ENST00000427810.2
ENST00000005082.9 ENST00000534569.1 ENST00000438262.2 ENST00000528796.1 ENST00000528410.1 ENST00000529678.1 ENST00000354599.6 ENST00000526601.1 ENST00000525502.1 ENST00000533036.1 ENST00000399602.4 |
ZNF195
|
zinc finger protein 195 |
| chr1_+_24645807 | 0.23 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr14_+_97925151 | 0.23 |
ENST00000554862.1
ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1
|
CTD-2506J14.1 |
| chr2_+_11864458 | 0.23 |
ENST00000396098.1
ENST00000396099.1 ENST00000425416.2 |
LPIN1
|
lipin 1 |
| chr2_-_224467093 | 0.23 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr12_-_13248562 | 0.22 |
ENST00000457134.2
ENST00000537302.1 |
GSG1
|
germ cell associated 1 |
| chr5_-_43515231 | 0.22 |
ENST00000306862.2
|
C5orf34
|
chromosome 5 open reading frame 34 |
| chr1_-_11024258 | 0.22 |
ENST00000418570.2
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chr11_-_3400442 | 0.22 |
ENST00000429541.2
ENST00000532539.1 |
ZNF195
|
zinc finger protein 195 |
| chr12_+_122880045 | 0.22 |
ENST00000539034.1
ENST00000535976.1 |
RP11-450K4.1
|
RP11-450K4.1 |
| chr19_+_50528971 | 0.21 |
ENST00000598809.1
ENST00000595661.1 ENST00000391821.2 |
ZNF473
|
zinc finger protein 473 |
| chr7_+_117251671 | 0.21 |
ENST00000468795.1
|
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr14_-_78174344 | 0.21 |
ENST00000216489.3
|
ALKBH1
|
alkB, alkylation repair homolog 1 (E. coli) |
| chr6_-_133035185 | 0.21 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr12_-_54778471 | 0.21 |
ENST00000550120.1
ENST00000394313.2 ENST00000547210.1 |
ZNF385A
|
zinc finger protein 385A |
| chr7_+_23749767 | 0.21 |
ENST00000355870.3
|
STK31
|
serine/threonine kinase 31 |
| chr3_+_178276488 | 0.21 |
ENST00000432997.1
ENST00000455865.1 |
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr2_+_233390863 | 0.21 |
ENST00000449596.1
ENST00000543200.1 |
CHRND
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr15_+_33022885 | 0.21 |
ENST00000322805.4
|
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chrX_-_13835147 | 0.21 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr3_+_63428752 | 0.21 |
ENST00000295894.5
|
SYNPR
|
synaptoporin |
| chr5_+_149877334 | 0.21 |
ENST00000523767.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr12_-_3982548 | 0.21 |
ENST00000397096.2
ENST00000447133.3 ENST00000450737.2 |
PARP11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr3_+_157154578 | 0.20 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr5_-_148758839 | 0.20 |
ENST00000261796.3
|
IL17B
|
interleukin 17B |
| chr3_+_138340067 | 0.20 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr12_-_9913489 | 0.20 |
ENST00000228434.3
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr4_-_77328458 | 0.20 |
ENST00000388914.3
ENST00000434846.2 |
CCDC158
|
coiled-coil domain containing 158 |
| chr10_+_64893039 | 0.20 |
ENST00000277746.6
ENST00000435510.2 |
NRBF2
|
nuclear receptor binding factor 2 |
| chr2_+_32502952 | 0.20 |
ENST00000238831.4
|
YIPF4
|
Yip1 domain family, member 4 |
| chr15_+_66797455 | 0.19 |
ENST00000446801.2
|
ZWILCH
|
zwilch kinetochore protein |
| chr7_+_54609995 | 0.19 |
ENST00000302287.3
ENST00000407838.3 |
VSTM2A
|
V-set and transmembrane domain containing 2A |
| chr13_-_46425865 | 0.19 |
ENST00000400405.2
|
SIAH3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr4_+_70796784 | 0.19 |
ENST00000246891.4
ENST00000444405.3 |
CSN1S1
|
casein alpha s1 |
| chr16_-_18911366 | 0.19 |
ENST00000565224.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr1_-_207095324 | 0.19 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr1_+_101702417 | 0.19 |
ENST00000305352.6
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr9_+_71819927 | 0.19 |
ENST00000535702.1
|
TJP2
|
tight junction protein 2 |
| chr7_+_23749945 | 0.19 |
ENST00000354639.3
ENST00000531170.1 ENST00000444333.2 ENST00000428484.1 |
STK31
|
serine/threonine kinase 31 |
| chr5_-_54281491 | 0.18 |
ENST00000381405.4
|
ESM1
|
endothelial cell-specific molecule 1 |
| chr11_-_108464321 | 0.18 |
ENST00000265843.4
|
EXPH5
|
exophilin 5 |
| chr1_+_104615595 | 0.18 |
ENST00000418362.1
|
RP11-364B6.1
|
RP11-364B6.1 |
| chr9_-_27529726 | 0.18 |
ENST00000262244.5
|
MOB3B
|
MOB kinase activator 3B |
| chr3_-_48130707 | 0.18 |
ENST00000360240.6
ENST00000383737.4 |
MAP4
|
microtubule-associated protein 4 |
| chr8_+_55528627 | 0.18 |
ENST00000220676.1
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant) |
| chr1_+_192127578 | 0.18 |
ENST00000367460.3
|
RGS18
|
regulator of G-protein signaling 18 |
| chr17_+_73663470 | 0.17 |
ENST00000583536.1
|
SAP30BP
|
SAP30 binding protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.7 | 4.6 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.6 | 3.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.5 | 1.5 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.5 | 1.8 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.3 | 1.9 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.3 | 3.0 | GO:1901509 | positive regulation of cAMP-mediated signaling(GO:0043950) regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.3 | 1.3 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.3 | 1.5 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.3 | 1.3 | GO:0048880 | sensory system development(GO:0048880) |
| 0.2 | 3.1 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.2 | 1.5 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.2 | 0.7 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.2 | 0.7 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 1.7 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 0.5 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.4 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 0.3 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.3 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.6 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.1 | 0.7 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.1 | 0.4 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.5 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 2.0 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.7 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.1 | 0.7 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 1.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 1.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.5 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.1 | 0.2 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.1 | 0.2 | GO:0035552 | tRNA wobble cytosine modification(GO:0002101) oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.1 | 0.2 | GO:0090291 | negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 0.4 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.2 | GO:0070352 | positive regulation of white fat cell proliferation(GO:0070352) |
| 0.1 | 2.0 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 0.2 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.1 | 0.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.2 | GO:0048627 | myoblast development(GO:0048627) |
| 0.1 | 0.3 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.1 | 0.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.2 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.2 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 1.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.5 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.2 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.0 | 0.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.0 | 0.1 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.2 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.3 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.3 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.2 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.1 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.4 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.3 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.2 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.5 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 1.0 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0050757 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.2 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 1.0 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.9 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.4 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.3 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.4 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.2 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.2 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.6 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.5 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.8 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.7 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.2 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.2 | GO:0033152 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.2 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.3 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 1.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.2 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 1.3 | GO:0030071 | regulation of mitotic metaphase/anaphase transition(GO:0030071) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.7 | GO:0042461 | photoreceptor cell development(GO:0042461) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.1 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:1903764 | cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.0 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.0 | GO:1990637 | response to prolactin(GO:1990637) regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.3 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.4 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.3 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.3 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.6 | 1.8 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.4 | 3.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 1.5 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.1 | 0.4 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 0.8 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.3 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 0.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.3 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 0.4 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 1.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.3 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 2.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 1.0 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 1.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.1 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 1.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 2.8 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 3.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.9 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.6 | 3.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.6 | 3.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.6 | 4.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.5 | 1.5 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.4 | 1.3 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.4 | 1.8 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.3 | 1.9 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 2.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 2.0 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 0.8 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 0.6 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.7 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.9 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.4 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.3 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.2 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.2 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 0.3 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.3 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 0.2 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 0.4 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.5 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 2.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.3 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.6 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.5 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.5 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 1.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 1.0 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.2 | GO:0030614 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 0.2 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 1.1 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.2 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0070016 | gamma-catenin binding(GO:0045295) armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 1.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 3.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 3.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 0.8 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 3.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 3.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 1.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 3.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 3.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 3.5 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.1 | 6.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 3.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.1 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |