Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for HOXB3
Z-value: 0.52
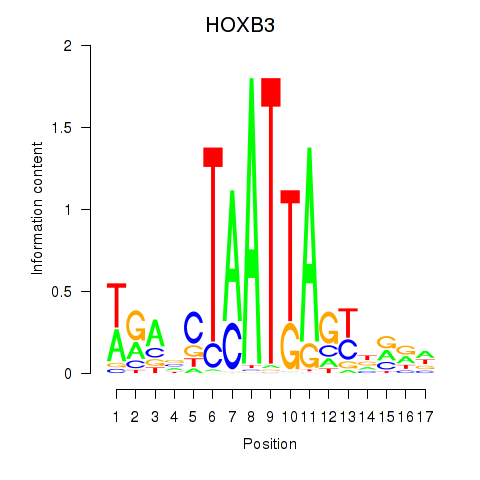
Transcription factors associated with HOXB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB3
|
ENSG00000120093.7 | homeobox B3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB3 | hg19_v2_chr17_-_46667628_46667642 | -0.12 | 5.7e-01 | Click! |
Activity profile of HOXB3 motif
Sorted Z-values of HOXB3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_102651343 | 1.20 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr6_+_127898312 | 0.91 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr4_+_86525299 | 0.87 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr5_-_61031495 | 0.68 |
ENST00000506550.1
ENST00000512882.2 |
CTD-2170G1.2
|
CTD-2170G1.2 |
| chr1_+_79115503 | 0.68 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr6_-_82462425 | 0.62 |
ENST00000369754.3
ENST00000320172.6 ENST00000369756.3 |
FAM46A
|
family with sequence similarity 46, member A |
| chr7_-_92777606 | 0.54 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr21_-_37914898 | 0.48 |
ENST00000399136.1
|
CLDN14
|
claudin 14 |
| chr11_+_5710919 | 0.46 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr1_+_62439037 | 0.45 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr2_-_202563414 | 0.44 |
ENST00000409474.3
ENST00000315506.7 ENST00000359962.5 |
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr1_+_160160346 | 0.43 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr12_-_51422017 | 0.42 |
ENST00000394904.3
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr1_+_160160283 | 0.42 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr15_-_55562479 | 0.41 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr2_+_143635067 | 0.39 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr15_-_55563072 | 0.39 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr18_+_61554932 | 0.39 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr11_-_117748138 | 0.38 |
ENST00000527717.1
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr11_-_104905840 | 0.37 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr18_+_55888767 | 0.37 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr15_-_55562582 | 0.36 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr11_+_33061543 | 0.35 |
ENST00000432887.1
ENST00000528898.1 ENST00000531632.2 |
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr2_-_220264703 | 0.35 |
ENST00000519905.1
ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr5_-_159546396 | 0.34 |
ENST00000523662.1
ENST00000456329.3 ENST00000307063.7 |
PWWP2A
|
PWWP domain containing 2A |
| chr2_-_120282070 | 0.33 |
ENST00000019103.5
|
SCTR
|
secretin receptor |
| chr1_-_67266939 | 0.32 |
ENST00000304526.2
|
INSL5
|
insulin-like 5 |
| chr19_-_3557570 | 0.31 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr15_-_101835110 | 0.31 |
ENST00000560496.1
|
SNRPA1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr2_-_10587897 | 0.30 |
ENST00000405333.1
ENST00000443218.1 |
ODC1
|
ornithine decarboxylase 1 |
| chr10_-_31288398 | 0.30 |
ENST00000538351.2
|
ZNF438
|
zinc finger protein 438 |
| chr7_-_99764853 | 0.30 |
ENST00000411994.1
ENST00000426974.2 |
GAL3ST4
|
galactose-3-O-sulfotransferase 4 |
| chrX_+_144908928 | 0.30 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr5_+_110427983 | 0.30 |
ENST00000513710.2
ENST00000505303.1 |
WDR36
|
WD repeat domain 36 |
| chr16_+_50730910 | 0.29 |
ENST00000300589.2
|
NOD2
|
nucleotide-binding oligomerization domain containing 2 |
| chr22_-_32651326 | 0.28 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr10_-_22292613 | 0.28 |
ENST00000376980.3
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr12_+_20848377 | 0.28 |
ENST00000540354.1
ENST00000266509.2 ENST00000381552.1 |
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr22_+_21996549 | 0.27 |
ENST00000248958.4
|
SDF2L1
|
stromal cell-derived factor 2-like 1 |
| chr10_-_4285923 | 0.27 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr4_-_120243545 | 0.27 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr1_+_144989309 | 0.26 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr6_-_62996066 | 0.26 |
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr2_+_187371440 | 0.25 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr16_+_31885079 | 0.25 |
ENST00000300870.10
ENST00000394846.3 |
ZNF267
|
zinc finger protein 267 |
| chr2_+_11682790 | 0.24 |
ENST00000389825.3
ENST00000381483.2 |
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr2_+_143635222 | 0.24 |
ENST00000375773.2
ENST00000409512.1 ENST00000410015.2 |
KYNU
|
kynureninase |
| chr12_+_112563335 | 0.23 |
ENST00000549358.1
ENST00000257604.5 ENST00000548092.1 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr12_+_112563303 | 0.23 |
ENST00000412615.2
|
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr7_+_120628731 | 0.22 |
ENST00000310396.5
|
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr11_-_107729887 | 0.22 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr12_-_10959892 | 0.22 |
ENST00000240615.2
|
TAS2R8
|
taste receptor, type 2, member 8 |
| chr9_-_5339873 | 0.22 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr6_+_53883708 | 0.21 |
ENST00000514921.1
ENST00000274897.5 ENST00000370877.2 |
MLIP
|
muscular LMNA-interacting protein |
| chr2_+_183982238 | 0.21 |
ENST00000442895.2
ENST00000446612.1 ENST00000409798.1 |
NUP35
|
nucleoporin 35kDa |
| chr11_-_104827425 | 0.21 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr2_+_65215604 | 0.21 |
ENST00000531327.1
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr1_+_160370344 | 0.21 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr16_-_28621298 | 0.21 |
ENST00000566189.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr12_+_28410128 | 0.21 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr20_+_9494987 | 0.20 |
ENST00000427562.2
ENST00000246070.2 |
LAMP5
|
lysosomal-associated membrane protein family, member 5 |
| chr8_-_13134045 | 0.20 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr15_+_64680003 | 0.20 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr13_-_47471155 | 0.20 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr19_+_10362577 | 0.20 |
ENST00000592514.1
ENST00000307422.5 ENST00000253099.6 ENST00000590150.1 ENST00000590669.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr13_+_102142296 | 0.20 |
ENST00000376162.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr11_-_26593779 | 0.19 |
ENST00000529533.1
|
MUC15
|
mucin 15, cell surface associated |
| chr3_+_147795932 | 0.19 |
ENST00000490465.1
|
RP11-639B1.1
|
RP11-639B1.1 |
| chr10_+_18549645 | 0.19 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr16_-_28621312 | 0.19 |
ENST00000314752.7
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr1_+_155006300 | 0.19 |
ENST00000295542.1
ENST00000392480.1 ENST00000423025.2 ENST00000368419.2 |
DCST1
|
DC-STAMP domain containing 1 |
| chr7_-_24797546 | 0.19 |
ENST00000414428.1
ENST00000419307.1 ENST00000342947.3 |
DFNA5
|
deafness, autosomal dominant 5 |
| chr19_+_10362882 | 0.19 |
ENST00000393733.2
ENST00000588502.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr2_+_196313239 | 0.19 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chrX_-_48824793 | 0.18 |
ENST00000376477.1
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr1_-_231004220 | 0.18 |
ENST00000366663.5
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr10_+_6779326 | 0.18 |
ENST00000417112.1
|
RP11-554I8.2
|
RP11-554I8.2 |
| chr12_-_122985067 | 0.18 |
ENST00000540586.1
ENST00000543897.1 |
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chrX_-_77225135 | 0.18 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr5_-_96478466 | 0.17 |
ENST00000274382.4
|
LIX1
|
Lix1 homolog (chicken) |
| chr19_+_48949030 | 0.17 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr9_+_130911770 | 0.17 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr2_+_208423840 | 0.17 |
ENST00000539789.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr15_-_101835414 | 0.17 |
ENST00000254193.6
|
SNRPA1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr14_+_74034310 | 0.16 |
ENST00000538782.1
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr3_-_151034734 | 0.16 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr16_-_31085514 | 0.16 |
ENST00000300849.4
|
ZNF668
|
zinc finger protein 668 |
| chr11_-_26593649 | 0.16 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr7_+_5919458 | 0.16 |
ENST00000416608.1
|
OCM
|
oncomodulin |
| chr12_+_7014126 | 0.16 |
ENST00000415834.1
ENST00000436789.1 |
LRRC23
|
leucine rich repeat containing 23 |
| chr11_-_26593677 | 0.16 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr10_+_53806501 | 0.16 |
ENST00000373975.2
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr4_+_110736659 | 0.15 |
ENST00000394631.3
ENST00000226796.6 |
GAR1
|
GAR1 ribonucleoprotein |
| chr2_+_29336855 | 0.15 |
ENST00000404424.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chrX_-_39923656 | 0.15 |
ENST00000413905.1
|
BCOR
|
BCL6 corepressor |
| chr17_-_73901494 | 0.15 |
ENST00000309352.3
|
MRPL38
|
mitochondrial ribosomal protein L38 |
| chr7_+_129015484 | 0.15 |
ENST00000490911.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr1_-_51796226 | 0.15 |
ENST00000451380.1
ENST00000371747.3 ENST00000439482.1 |
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr2_+_168675182 | 0.14 |
ENST00000305861.1
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr16_-_11036300 | 0.14 |
ENST00000331808.4
|
DEXI
|
Dexi homolog (mouse) |
| chr19_-_6737576 | 0.14 |
ENST00000601716.1
ENST00000264080.7 |
GPR108
|
G protein-coupled receptor 108 |
| chr14_+_22320634 | 0.14 |
ENST00000390435.1
|
TRAV8-3
|
T cell receptor alpha variable 8-3 |
| chr2_-_89160770 | 0.14 |
ENST00000390240.2
|
IGKJ3
|
immunoglobulin kappa joining 3 |
| chr11_-_104817919 | 0.14 |
ENST00000533252.1
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr20_-_5485166 | 0.14 |
ENST00000589201.1
ENST00000379053.4 |
LINC00654
|
long intergenic non-protein coding RNA 654 |
| chr7_+_99202003 | 0.14 |
ENST00000609449.1
|
GS1-259H13.2
|
GS1-259H13.2 |
| chr6_+_26045603 | 0.13 |
ENST00000540144.1
|
HIST1H3C
|
histone cluster 1, H3c |
| chr17_-_40337470 | 0.13 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr19_-_46088068 | 0.12 |
ENST00000263275.4
ENST00000323060.3 |
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr10_-_27529486 | 0.12 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr17_-_8770956 | 0.12 |
ENST00000311434.9
|
PIK3R6
|
phosphoinositide-3-kinase, regulatory subunit 6 |
| chr9_+_130911723 | 0.12 |
ENST00000277480.2
ENST00000373013.2 ENST00000540948.1 |
LCN2
|
lipocalin 2 |
| chr4_+_113568207 | 0.12 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr4_-_164534657 | 0.11 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr7_-_121944491 | 0.11 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr16_+_31366536 | 0.11 |
ENST00000562522.1
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr11_+_67219867 | 0.11 |
ENST00000438189.2
|
CABP4
|
calcium binding protein 4 |
| chr3_+_114012819 | 0.11 |
ENST00000383671.3
|
TIGIT
|
T cell immunoreceptor with Ig and ITIM domains |
| chr6_-_82957433 | 0.11 |
ENST00000306270.7
|
IBTK
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr2_-_88125471 | 0.11 |
ENST00000398146.3
|
RGPD2
|
RANBP2-like and GRIP domain containing 2 |
| chr14_+_57671888 | 0.11 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr7_-_122339162 | 0.10 |
ENST00000340112.2
|
RNF133
|
ring finger protein 133 |
| chr4_-_111563076 | 0.10 |
ENST00000354925.2
ENST00000511990.1 |
PITX2
|
paired-like homeodomain 2 |
| chr21_+_31768348 | 0.10 |
ENST00000355459.2
|
KRTAP13-1
|
keratin associated protein 13-1 |
| chr12_-_5352315 | 0.10 |
ENST00000536518.1
|
RP11-319E16.1
|
RP11-319E16.1 |
| chr3_-_151047327 | 0.10 |
ENST00000325602.5
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13 |
| chr17_-_62208169 | 0.10 |
ENST00000606895.1
|
ERN1
|
endoplasmic reticulum to nucleus signaling 1 |
| chr12_+_22778116 | 0.10 |
ENST00000538218.1
|
ETNK1
|
ethanolamine kinase 1 |
| chr8_-_623547 | 0.10 |
ENST00000522893.1
|
ERICH1
|
glutamate-rich 1 |
| chrX_-_21676442 | 0.10 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr2_+_234545148 | 0.10 |
ENST00000373445.1
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr1_-_207226313 | 0.10 |
ENST00000367084.1
|
YOD1
|
YOD1 deubiquitinase |
| chr1_-_13390765 | 0.09 |
ENST00000357367.2
|
PRAMEF8
|
PRAME family member 8 |
| chr6_+_149721495 | 0.09 |
ENST00000326669.4
|
SUMO4
|
small ubiquitin-like modifier 4 |
| chr12_-_6740802 | 0.09 |
ENST00000431922.1
|
LPAR5
|
lysophosphatidic acid receptor 5 |
| chrX_+_41548259 | 0.09 |
ENST00000378138.5
|
GPR34
|
G protein-coupled receptor 34 |
| chr2_+_89184868 | 0.09 |
ENST00000390243.2
|
IGKV4-1
|
immunoglobulin kappa variable 4-1 |
| chr12_-_117175819 | 0.09 |
ENST00000261318.3
ENST00000536380.1 |
C12orf49
|
chromosome 12 open reading frame 49 |
| chr5_+_115177178 | 0.09 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr19_-_6424783 | 0.09 |
ENST00000398148.3
|
KHSRP
|
KH-type splicing regulatory protein |
| chr6_+_45296391 | 0.09 |
ENST00000371436.6
ENST00000576263.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr15_+_78830023 | 0.08 |
ENST00000599596.1
|
AC027228.1
|
HCG2005369; Uncharacterized protein |
| chr11_-_96076334 | 0.08 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr1_-_146040968 | 0.08 |
ENST00000401010.3
|
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr12_+_55248289 | 0.08 |
ENST00000308796.6
|
MUCL1
|
mucin-like 1 |
| chr5_+_129083772 | 0.08 |
ENST00000564719.1
|
KIAA1024L
|
KIAA1024-like |
| chrX_+_102024075 | 0.08 |
ENST00000431616.1
ENST00000440496.1 ENST00000420471.1 ENST00000435966.1 |
LINC00630
|
long intergenic non-protein coding RNA 630 |
| chr12_-_123187890 | 0.08 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chrX_+_41548220 | 0.08 |
ENST00000378142.4
|
GPR34
|
G protein-coupled receptor 34 |
| chr2_+_113816215 | 0.08 |
ENST00000346807.3
|
IL36RN
|
interleukin 36 receptor antagonist |
| chr4_-_8873531 | 0.08 |
ENST00000400677.3
|
HMX1
|
H6 family homeobox 1 |
| chr8_+_50824233 | 0.08 |
ENST00000522124.1
|
SNTG1
|
syntrophin, gamma 1 |
| chr12_-_123201337 | 0.08 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr2_-_113810444 | 0.08 |
ENST00000259213.4
ENST00000327407.2 |
IL36B
|
interleukin 36, beta |
| chr5_+_55149150 | 0.08 |
ENST00000297015.3
|
IL31RA
|
interleukin 31 receptor A |
| chr16_+_31724552 | 0.07 |
ENST00000539915.1
ENST00000316491.9 ENST00000399681.3 ENST00000398696.3 ENST00000534369.1 |
ZNF720
|
zinc finger protein 720 |
| chr11_-_119991589 | 0.07 |
ENST00000526881.1
|
TRIM29
|
tripartite motif containing 29 |
| chr1_+_197237352 | 0.07 |
ENST00000538660.1
ENST00000367400.3 ENST00000367399.2 |
CRB1
|
crumbs homolog 1 (Drosophila) |
| chr5_-_32444828 | 0.07 |
ENST00000265069.8
|
ZFR
|
zinc finger RNA binding protein |
| chr5_+_140019004 | 0.07 |
ENST00000394671.3
ENST00000511410.1 ENST00000537378.1 |
TMCO6
|
transmembrane and coiled-coil domains 6 |
| chrX_+_83116142 | 0.07 |
ENST00000329312.4
|
CYLC1
|
cylicin, basic protein of sperm head cytoskeleton 1 |
| chr4_-_138453559 | 0.07 |
ENST00000511115.1
|
PCDH18
|
protocadherin 18 |
| chr2_+_228736321 | 0.07 |
ENST00000309931.2
|
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr9_+_135937365 | 0.07 |
ENST00000372080.4
ENST00000351304.7 |
CEL
|
carboxyl ester lipase |
| chr18_+_268148 | 0.07 |
ENST00000581677.1
|
RP11-705O1.8
|
RP11-705O1.8 |
| chr1_+_202317815 | 0.07 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr1_+_156030937 | 0.07 |
ENST00000361084.5
|
RAB25
|
RAB25, member RAS oncogene family |
| chr14_-_24711865 | 0.07 |
ENST00000399423.4
ENST00000267415.7 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr12_-_78753496 | 0.07 |
ENST00000548512.1
|
RP11-38F22.1
|
RP11-38F22.1 |
| chr9_+_124329336 | 0.07 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr15_+_84841242 | 0.07 |
ENST00000558195.1
|
RP11-182J1.16
|
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr9_-_100707116 | 0.07 |
ENST00000259456.3
|
HEMGN
|
hemogen |
| chr14_-_24711806 | 0.07 |
ENST00000540705.1
ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr11_+_63273811 | 0.07 |
ENST00000340246.5
|
LGALS12
|
lectin, galactoside-binding, soluble, 12 |
| chr6_+_34204642 | 0.07 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr16_+_6069072 | 0.06 |
ENST00000547605.1
ENST00000550418.1 ENST00000553186.1 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr2_-_169887827 | 0.06 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr16_-_66864806 | 0.06 |
ENST00000566336.1
ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr2_+_197577841 | 0.06 |
ENST00000409270.1
|
CCDC150
|
coiled-coil domain containing 150 |
| chrX_+_106045891 | 0.06 |
ENST00000357242.5
ENST00000310452.2 ENST00000481617.2 ENST00000276175.3 |
TBC1D8B
|
TBC1 domain family, member 8B (with GRAM domain) |
| chr4_-_100212132 | 0.06 |
ENST00000209668.2
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chr6_+_26204825 | 0.06 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr1_+_207226574 | 0.06 |
ENST00000367080.3
ENST00000367079.2 |
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr3_-_119182523 | 0.06 |
ENST00000319172.5
|
TMEM39A
|
transmembrane protein 39A |
| chr6_-_119256279 | 0.06 |
ENST00000316068.3
|
MCM9
|
minichromosome maintenance complex component 9 |
| chr14_+_32798462 | 0.06 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr1_+_144173162 | 0.06 |
ENST00000356801.6
|
NBPF8
|
neuroblastoma breakpoint family, member 8 |
| chr3_-_126327398 | 0.06 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr2_-_3606206 | 0.06 |
ENST00000315212.3
|
RNASEH1
|
ribonuclease H1 |
| chr1_+_202317855 | 0.06 |
ENST00000356764.2
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr4_-_52883786 | 0.06 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr5_-_108063949 | 0.05 |
ENST00000606054.1
|
LINC01023
|
long intergenic non-protein coding RNA 1023 |
| chr6_+_36562132 | 0.05 |
ENST00000373715.6
ENST00000339436.7 |
SRSF3
|
serine/arginine-rich splicing factor 3 |
| chrX_+_135730373 | 0.05 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr6_-_133035185 | 0.05 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr11_-_95522907 | 0.05 |
ENST00000358780.5
ENST00000542135.1 |
FAM76B
|
family with sequence similarity 76, member B |
| chr10_-_135379132 | 0.05 |
ENST00000343131.5
|
SYCE1
|
synaptonemal complex central element protein 1 |
| chr5_+_140019079 | 0.05 |
ENST00000252100.6
|
TMCO6
|
transmembrane and coiled-coil domains 6 |
| chr4_+_40198527 | 0.05 |
ENST00000381799.5
|
RHOH
|
ras homolog family member H |
| chr2_+_166095898 | 0.05 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.2 | 1.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.3 | GO:0035419 | detection of peptidoglycan(GO:0032499) activation of MAPK activity involved in innate immune response(GO:0035419) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.1 | 0.4 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.4 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 0.8 | GO:0014877 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.3 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.2 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 0.3 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.2 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.2 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.3 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:0060578 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.2 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.2 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 1.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.0 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.0 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.4 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.3 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.2 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.0 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 0.4 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 1.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.7 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.2 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.2 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.4 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.2 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 1.0 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.5 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 1.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.2 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.2 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0030395 | lactose binding(GO:0030395) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.0 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.0 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |