Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for HOXB4_LHX9
Z-value: 0.56
Transcription factors associated with HOXB4_LHX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
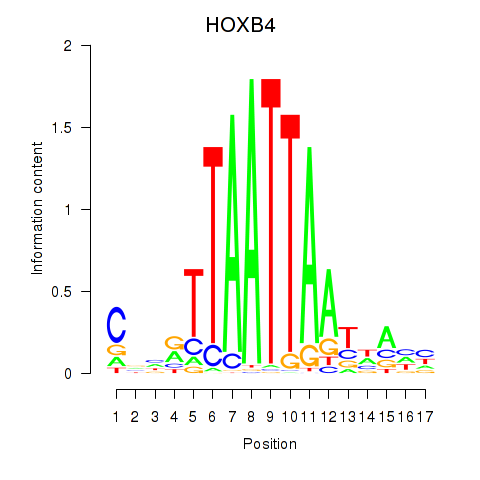
HOXB4
|
ENSG00000182742.5 | homeobox B4 |
|
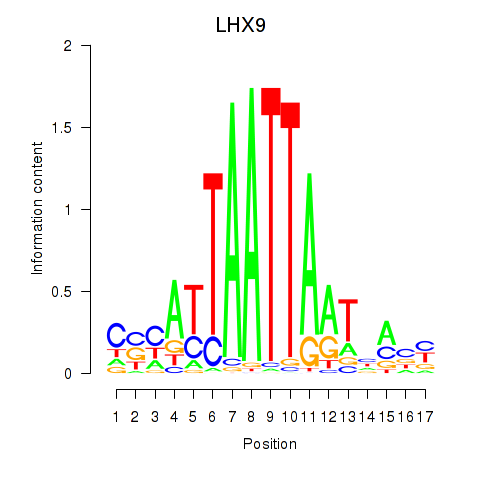
LHX9
|
ENSG00000143355.11 | LIM homeobox 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB4 | hg19_v2_chr17_-_46657473_46657473 | -0.40 | 4.5e-02 | Click! |
| LHX9 | hg19_v2_chr1_+_197881592_197881635 | 0.17 | 4.1e-01 | Click! |
Activity profile of HOXB4_LHX9 motif
Sorted Z-values of HOXB4_LHX9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_29527702 | 2.23 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr2_+_152214098 | 1.74 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr2_-_225811747 | 1.43 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr6_+_26402465 | 1.37 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr6_+_26440700 | 1.25 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr1_-_8000872 | 1.21 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr15_+_67418047 | 1.08 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr7_+_134528635 | 1.05 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr8_-_90996459 | 1.05 |
ENST00000517337.1
ENST00000409330.1 |
NBN
|
nibrin |
| chr2_+_102953608 | 0.98 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr15_+_64680003 | 0.96 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr8_-_90996837 | 0.96 |
ENST00000519426.1
ENST00000265433.3 |
NBN
|
nibrin |
| chr6_+_114178512 | 0.88 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr4_+_86525299 | 0.86 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr5_-_35938674 | 0.84 |
ENST00000397366.1
ENST00000513623.1 ENST00000514524.1 ENST00000397367.2 |
CAPSL
|
calcyphosine-like |
| chr6_-_32806506 | 0.81 |
ENST00000374897.2
ENST00000452392.2 |
TAP2
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr11_+_5710919 | 0.79 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr6_+_26402517 | 0.77 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr10_+_24755416 | 0.74 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr14_+_31046959 | 0.73 |
ENST00000547532.1
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr15_+_89631647 | 0.72 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr11_+_33061543 | 0.70 |
ENST00000432887.1
ENST00000528898.1 ENST00000531632.2 |
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr4_+_169418195 | 0.70 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr12_+_26348582 | 0.69 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr4_+_169418255 | 0.67 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr17_+_1674982 | 0.64 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr18_-_74839891 | 0.63 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr14_-_51027838 | 0.63 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr19_+_54466179 | 0.55 |
ENST00000270458.2
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr6_+_26365443 | 0.54 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr15_+_89631381 | 0.53 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chr9_-_26947453 | 0.52 |
ENST00000397292.3
|
PLAA
|
phospholipase A2-activating protein |
| chr4_-_103749205 | 0.51 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr3_-_157221380 | 0.51 |
ENST00000468233.1
|
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr22_+_46476192 | 0.51 |
ENST00000443490.1
|
FLJ27365
|
hsa-mir-4763 |
| chr5_-_16916624 | 0.50 |
ENST00000513882.1
|
MYO10
|
myosin X |
| chr11_-_107729887 | 0.50 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr1_-_57045228 | 0.49 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr3_-_191000172 | 0.47 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr6_+_29079668 | 0.45 |
ENST00000377169.1
|
OR2J3
|
olfactory receptor, family 2, subfamily J, member 3 |
| chr11_-_55703876 | 0.43 |
ENST00000301532.3
|
OR5I1
|
olfactory receptor, family 5, subfamily I, member 1 |
| chr20_+_31805131 | 0.43 |
ENST00000375454.3
ENST00000375452.3 |
BPIFA3
|
BPI fold containing family A, member 3 |
| chr17_+_66511540 | 0.43 |
ENST00000588188.2
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr8_+_50824233 | 0.43 |
ENST00000522124.1
|
SNTG1
|
syntrophin, gamma 1 |
| chr4_+_108745711 | 0.42 |
ENST00000394684.4
|
SGMS2
|
sphingomyelin synthase 2 |
| chr5_-_150473127 | 0.41 |
ENST00000521001.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr10_-_27529486 | 0.41 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr4_+_169552748 | 0.41 |
ENST00000504519.1
ENST00000512127.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr11_-_104827425 | 0.41 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr18_+_56806701 | 0.40 |
ENST00000587834.1
|
SEC11C
|
SEC11 homolog C (S. cerevisiae) |
| chr21_-_37914898 | 0.39 |
ENST00000399136.1
|
CLDN14
|
claudin 14 |
| chr8_+_104310661 | 0.39 |
ENST00000522566.1
|
FZD6
|
frizzled family receptor 6 |
| chr4_-_39979576 | 0.38 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr10_-_13043697 | 0.37 |
ENST00000378825.3
|
CCDC3
|
coiled-coil domain containing 3 |
| chr15_-_72523924 | 0.37 |
ENST00000566809.1
ENST00000567087.1 ENST00000569050.1 ENST00000568883.1 |
PKM
|
pyruvate kinase, muscle |
| chr8_-_116504448 | 0.36 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr1_+_225600404 | 0.36 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr4_-_103998439 | 0.36 |
ENST00000503230.1
ENST00000503818.1 |
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr13_+_97928395 | 0.36 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr19_-_3557570 | 0.36 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr3_-_157221357 | 0.34 |
ENST00000494677.1
|
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chrX_+_1710484 | 0.34 |
ENST00000313871.3
ENST00000381261.3 |
AKAP17A
|
A kinase (PRKA) anchor protein 17A |
| chr4_-_103749179 | 0.33 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_-_69536346 | 0.32 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr20_+_18488137 | 0.32 |
ENST00000450074.1
ENST00000262544.2 ENST00000336714.3 ENST00000377475.3 |
SEC23B
|
Sec23 homolog B (S. cerevisiae) |
| chrX_+_30261847 | 0.32 |
ENST00000378981.3
ENST00000397550.1 |
MAGEB1
|
melanoma antigen family B, 1 |
| chr4_+_119810134 | 0.31 |
ENST00000434046.2
|
SYNPO2
|
synaptopodin 2 |
| chr10_+_18549645 | 0.31 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr2_+_102456277 | 0.31 |
ENST00000421882.1
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr1_-_231005310 | 0.31 |
ENST00000470540.1
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr12_+_26348246 | 0.30 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr7_+_134430212 | 0.30 |
ENST00000436461.2
|
CALD1
|
caldesmon 1 |
| chr15_+_66679155 | 0.29 |
ENST00000307102.5
|
MAP2K1
|
mitogen-activated protein kinase kinase 1 |
| chr6_+_31553978 | 0.29 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr14_-_25078864 | 0.29 |
ENST00000216338.4
ENST00000557220.2 ENST00000382548.4 |
GZMH
|
granzyme H (cathepsin G-like 2, protein h-CCPX) |
| chr13_+_77522632 | 0.28 |
ENST00000377462.1
|
IRG1
|
immunoresponsive 1 homolog (mouse) |
| chrX_-_100662881 | 0.28 |
ENST00000218516.3
|
GLA
|
galactosidase, alpha |
| chr11_+_35201826 | 0.28 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr11_-_128894053 | 0.28 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr8_-_125577940 | 0.27 |
ENST00000519168.1
ENST00000395508.2 |
MTSS1
|
metastasis suppressor 1 |
| chr2_+_187454749 | 0.27 |
ENST00000261023.3
ENST00000374907.3 |
ITGAV
|
integrin, alpha V |
| chr12_+_112279782 | 0.27 |
ENST00000550735.2
|
MAPKAPK5
|
mitogen-activated protein kinase-activated protein kinase 5 |
| chr8_-_30670384 | 0.27 |
ENST00000221138.4
ENST00000518243.1 |
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr12_+_26348429 | 0.27 |
ENST00000242729.2
|
SSPN
|
sarcospan |
| chr4_-_120243545 | 0.26 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chrX_+_108779004 | 0.26 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr14_-_45252031 | 0.26 |
ENST00000556405.1
|
RP11-398E10.1
|
RP11-398E10.1 |
| chr10_-_75415825 | 0.25 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr7_-_140482926 | 0.25 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr14_+_103851712 | 0.25 |
ENST00000440884.3
ENST00000416682.2 ENST00000429436.2 ENST00000303622.9 |
MARK3
|
MAP/microtubule affinity-regulating kinase 3 |
| chr19_-_14945933 | 0.24 |
ENST00000322301.3
|
OR7A5
|
olfactory receptor, family 7, subfamily A, member 5 |
| chr11_-_119991589 | 0.24 |
ENST00000526881.1
|
TRIM29
|
tripartite motif containing 29 |
| chr4_-_119759795 | 0.24 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr7_-_87856303 | 0.23 |
ENST00000394641.3
|
SRI
|
sorcin |
| chr6_-_89927151 | 0.23 |
ENST00000454853.2
|
GABRR1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chrX_-_15332665 | 0.23 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr2_-_169887827 | 0.23 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr7_-_87856280 | 0.22 |
ENST00000490437.1
ENST00000431660.1 |
SRI
|
sorcin |
| chr4_-_103998060 | 0.22 |
ENST00000339611.4
|
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr7_-_25702669 | 0.22 |
ENST00000446840.1
|
AC003090.1
|
AC003090.1 |
| chr2_-_136678123 | 0.21 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr14_+_22465771 | 0.21 |
ENST00000390445.2
|
TRAV17
|
T cell receptor alpha variable 17 |
| chr20_-_1447467 | 0.21 |
ENST00000353088.2
ENST00000350991.4 |
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr16_-_29934558 | 0.21 |
ENST00000568995.1
ENST00000566413.1 |
KCTD13
|
potassium channel tetramerization domain containing 13 |
| chr1_+_160370344 | 0.20 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr20_-_1447547 | 0.20 |
ENST00000476071.1
|
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr18_-_51750948 | 0.20 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr22_-_32651326 | 0.20 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr3_-_122233723 | 0.20 |
ENST00000493510.1
ENST00000344337.6 ENST00000476916.1 ENST00000465882.1 |
KPNA1
|
karyopherin alpha 1 (importin alpha 5) |
| chr21_+_33671160 | 0.20 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr3_-_141747950 | 0.20 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr6_-_87804815 | 0.19 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr3_+_111718173 | 0.19 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chrX_+_44703249 | 0.19 |
ENST00000339042.4
|
DUSP21
|
dual specificity phosphatase 21 |
| chrX_-_77225135 | 0.19 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr11_-_121986923 | 0.19 |
ENST00000560104.1
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr4_-_103997862 | 0.19 |
ENST00000394785.3
|
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr6_+_42584847 | 0.18 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr2_-_128284020 | 0.18 |
ENST00000295321.4
ENST00000455721.2 |
IWS1
|
IWS1 homolog (S. cerevisiae) |
| chr3_+_138340049 | 0.18 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr4_-_103749105 | 0.18 |
ENST00000394801.4
ENST00000394804.2 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr10_+_35415851 | 0.18 |
ENST00000374726.3
|
CREM
|
cAMP responsive element modulator |
| chr10_+_90424196 | 0.18 |
ENST00000394375.3
ENST00000608620.1 ENST00000238983.4 ENST00000355843.2 |
LIPF
|
lipase, gastric |
| chr2_+_90248739 | 0.17 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr20_-_56265680 | 0.17 |
ENST00000414037.1
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr14_+_95027772 | 0.17 |
ENST00000555095.1
ENST00000298841.5 ENST00000554220.1 ENST00000553780.1 |
SERPINA4
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4 serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr14_-_53258314 | 0.17 |
ENST00000216410.3
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr14_-_72458326 | 0.17 |
ENST00000542853.1
|
AC005477.1
|
AC005477.1 |
| chr4_-_39033963 | 0.17 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr4_-_10686373 | 0.17 |
ENST00000442825.2
|
CLNK
|
cytokine-dependent hematopoietic cell linker |
| chrX_+_83116142 | 0.17 |
ENST00000329312.4
|
CYLC1
|
cylicin, basic protein of sperm head cytoskeleton 1 |
| chr18_-_3219847 | 0.16 |
ENST00000261606.7
|
MYOM1
|
myomesin 1 |
| chr20_+_30697298 | 0.16 |
ENST00000398022.2
|
TM9SF4
|
transmembrane 9 superfamily protein member 4 |
| chr12_-_22063787 | 0.16 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr12_-_23737534 | 0.16 |
ENST00000396007.2
|
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr5_-_172036436 | 0.15 |
ENST00000601856.1
|
AC027309.1
|
AC027309.1 |
| chr15_+_89922345 | 0.15 |
ENST00000558982.1
|
LINC00925
|
long intergenic non-protein coding RNA 925 |
| chr1_+_157963063 | 0.15 |
ENST00000360089.4
ENST00000368173.3 ENST00000392272.2 |
KIRREL
|
kin of IRRE like (Drosophila) |
| chr11_+_60383204 | 0.15 |
ENST00000412599.1
ENST00000320202.4 |
LINC00301
|
long intergenic non-protein coding RNA 301 |
| chr13_+_32313658 | 0.15 |
ENST00000380314.1
ENST00000298386.2 |
RXFP2
|
relaxin/insulin-like family peptide receptor 2 |
| chr1_+_28261492 | 0.14 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr4_+_100432161 | 0.14 |
ENST00000326581.4
ENST00000514652.1 |
C4orf17
|
chromosome 4 open reading frame 17 |
| chr1_+_196743912 | 0.14 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr5_+_66300446 | 0.14 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr17_-_72772462 | 0.14 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chrX_-_21676442 | 0.14 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr11_-_101000445 | 0.14 |
ENST00000534013.1
|
PGR
|
progesterone receptor |
| chrX_+_36254051 | 0.13 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr2_-_161056802 | 0.13 |
ENST00000283249.2
ENST00000409872.1 |
ITGB6
|
integrin, beta 6 |
| chr4_-_185275104 | 0.13 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr17_-_39324424 | 0.13 |
ENST00000391356.2
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr4_-_103748880 | 0.13 |
ENST00000453744.2
ENST00000349311.8 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_-_103749313 | 0.13 |
ENST00000394803.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr7_-_108168580 | 0.13 |
ENST00000453085.1
|
PNPLA8
|
patatin-like phospholipase domain containing 8 |
| chr6_-_91296737 | 0.13 |
ENST00000369332.3
ENST00000369329.3 |
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr6_-_91296602 | 0.13 |
ENST00000369325.3
ENST00000369327.3 |
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr20_-_21494654 | 0.13 |
ENST00000377142.4
|
NKX2-2
|
NK2 homeobox 2 |
| chr6_+_36562132 | 0.13 |
ENST00000373715.6
ENST00000339436.7 |
SRSF3
|
serine/arginine-rich splicing factor 3 |
| chr3_+_38537960 | 0.13 |
ENST00000453767.1
|
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr9_-_28670283 | 0.13 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr17_-_4938712 | 0.13 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr7_-_111032971 | 0.12 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr21_-_31538971 | 0.12 |
ENST00000286808.3
|
CLDN17
|
claudin 17 |
| chr16_+_31271274 | 0.12 |
ENST00000287497.8
ENST00000544665.3 |
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr14_-_80697396 | 0.12 |
ENST00000557010.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr7_+_120628731 | 0.12 |
ENST00000310396.5
|
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr1_-_153518270 | 0.12 |
ENST00000354332.4
ENST00000368716.4 |
S100A4
|
S100 calcium binding protein A4 |
| chr19_-_41870026 | 0.12 |
ENST00000243578.3
|
B9D2
|
B9 protein domain 2 |
| chr17_-_45266542 | 0.12 |
ENST00000531206.1
ENST00000527547.1 ENST00000446365.2 ENST00000575483.1 ENST00000066544.3 |
CDC27
|
cell division cycle 27 |
| chr3_-_58613323 | 0.12 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr1_-_152131703 | 0.12 |
ENST00000316073.3
|
RPTN
|
repetin |
| chrX_+_19362011 | 0.12 |
ENST00000379806.5
ENST00000545074.1 ENST00000540249.1 ENST00000423505.1 ENST00000417819.1 ENST00000422285.2 ENST00000355808.5 ENST00000379805.3 |
PDHA1
|
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr1_-_47184745 | 0.12 |
ENST00000544071.1
|
EFCAB14
|
EF-hand calcium binding domain 14 |
| chr21_-_42219065 | 0.12 |
ENST00000400454.1
|
DSCAM
|
Down syndrome cell adhesion molecule |
| chr3_+_139063372 | 0.12 |
ENST00000478464.1
|
MRPS22
|
mitochondrial ribosomal protein S22 |
| chr18_+_21033239 | 0.12 |
ENST00000581585.1
ENST00000577501.1 |
RIOK3
|
RIO kinase 3 |
| chr11_-_75917569 | 0.12 |
ENST00000322563.3
|
WNT11
|
wingless-type MMTV integration site family, member 11 |
| chr8_+_105235572 | 0.12 |
ENST00000523362.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr7_-_86849883 | 0.11 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr14_+_56584414 | 0.11 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr8_+_9953214 | 0.11 |
ENST00000382490.5
|
MSRA
|
methionine sulfoxide reductase A |
| chr3_+_111717511 | 0.11 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr11_+_55594695 | 0.11 |
ENST00000378397.1
|
OR5L2
|
olfactory receptor, family 5, subfamily L, member 2 |
| chr3_+_138340067 | 0.11 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr8_+_9953061 | 0.11 |
ENST00000522907.1
ENST00000528246.1 |
MSRA
|
methionine sulfoxide reductase A |
| chr5_-_159546396 | 0.11 |
ENST00000523662.1
ENST00000456329.3 ENST00000307063.7 |
PWWP2A
|
PWWP domain containing 2A |
| chr1_-_89357179 | 0.11 |
ENST00000448623.1
ENST00000418217.1 ENST00000370500.5 |
GTF2B
|
general transcription factor IIB |
| chr8_-_90769422 | 0.11 |
ENST00000524190.1
ENST00000523859.1 |
RP11-37B2.1
|
RP11-37B2.1 |
| chr4_+_119809984 | 0.11 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr3_+_111717600 | 0.11 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr4_-_159956333 | 0.11 |
ENST00000434826.2
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr6_+_47624172 | 0.11 |
ENST00000507065.1
ENST00000296862.1 |
GPR111
|
G protein-coupled receptor 111 |
| chr2_+_27255806 | 0.11 |
ENST00000238788.9
ENST00000404032.3 |
TMEM214
|
transmembrane protein 214 |
| chr14_-_107049312 | 0.10 |
ENST00000390627.2
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr1_+_12851545 | 0.10 |
ENST00000332296.7
|
PRAMEF1
|
PRAME family member 1 |
| chr1_-_205091115 | 0.10 |
ENST00000264515.6
ENST00000367164.1 |
RBBP5
|
retinoblastoma binding protein 5 |
| chr14_+_22670455 | 0.10 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr12_-_52604607 | 0.10 |
ENST00000551894.1
ENST00000553017.1 |
C12orf80
|
chromosome 12 open reading frame 80 |
| chr15_+_69857515 | 0.10 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr4_+_72897521 | 0.10 |
ENST00000308744.6
ENST00000344413.5 |
NPFFR2
|
neuropeptide FF receptor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB4_LHX9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.4 | 2.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.3 | 0.8 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.2 | 1.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.2 | 1.2 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.2 | 1.0 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 2.7 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.1 | 0.1 | GO:0060031 | mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.1 | 0.6 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 0.4 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 1.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.4 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 0.8 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.6 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 1.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.2 | GO:0061346 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 0.3 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.1 | 2.0 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.5 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.3 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.3 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.3 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.4 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 1.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.4 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.3 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.2 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.5 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.3 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.3 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.2 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.2 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 1.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.4 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 | 0.1 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.1 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 0.2 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 1.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.0 | GO:2000053 | negative regulation of B cell differentiation(GO:0045578) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.0 | 0.3 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.2 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.1 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.0 | 0.0 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.2 | 2.0 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.4 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 1.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.8 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.4 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 1.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 1.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.5 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.2 | GO:0036030 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.1 | 0.2 | GO:0033150 | cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.1 | 0.6 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 1.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.3 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 2.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.0 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 1.8 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 1.8 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.0 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 1.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.2 | 1.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.2 | 0.8 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.2 | 0.8 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.1 | 2.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.4 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 2.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 1.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.2 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 2.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.5 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.2 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 1.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 1.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.5 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.5 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 1.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.2 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.1 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 0.9 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 2.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.5 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |