Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
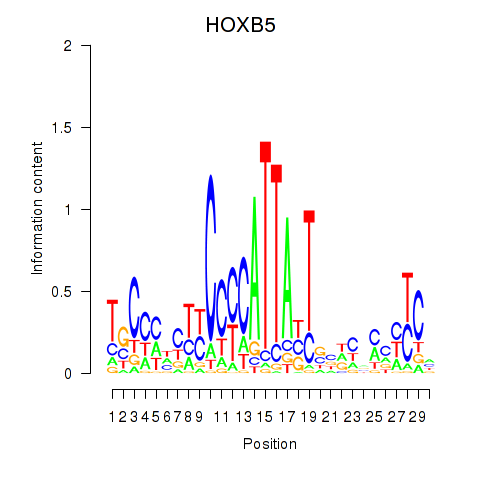
Results for HOXB5
Z-value: 0.43
Transcription factors associated with HOXB5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB5
|
ENSG00000120075.5 | homeobox B5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB5 | hg19_v2_chr17_-_46671323_46671323 | -0.47 | 1.9e-02 | Click! |
Activity profile of HOXB5 motif
Sorted Z-values of HOXB5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_127541194 | 2.54 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase |
| chr6_-_32820529 | 1.67 |
ENST00000425148.2
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr3_-_164914640 | 0.71 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK-like family, member 3 |
| chr16_-_84651647 | 0.70 |
ENST00000564057.1
|
COTL1
|
coactosin-like 1 (Dictyostelium) |
| chr15_-_72523454 | 0.66 |
ENST00000565154.1
ENST00000565184.1 ENST00000389093.3 ENST00000449901.2 ENST00000335181.5 ENST00000319622.6 |
PKM
|
pyruvate kinase, muscle |
| chr1_+_146373546 | 0.65 |
ENST00000446760.2
|
NBPF12
|
neuroblastoma breakpoint family, member 12 |
| chr5_-_35048047 | 0.55 |
ENST00000231420.6
|
AGXT2
|
alanine--glyoxylate aminotransferase 2 |
| chr4_-_114900831 | 0.55 |
ENST00000315366.7
|
ARSJ
|
arylsulfatase family, member J |
| chr1_+_155579979 | 0.49 |
ENST00000452804.2
ENST00000538143.1 ENST00000245564.2 ENST00000368341.4 |
MSTO1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr11_-_57089671 | 0.48 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr1_+_22307592 | 0.47 |
ENST00000400277.2
|
CELA3B
|
chymotrypsin-like elastase family, member 3B |
| chr10_+_114135952 | 0.43 |
ENST00000356116.1
ENST00000433418.1 ENST00000354273.4 |
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr5_+_133842243 | 0.37 |
ENST00000515627.2
|
AC005355.2
|
AC005355.2 |
| chr14_-_24020858 | 0.36 |
ENST00000419474.3
|
ZFHX2
|
zinc finger homeobox 2 |
| chr1_+_4714792 | 0.33 |
ENST00000378190.3
|
AJAP1
|
adherens junctions associated protein 1 |
| chr16_-_1821496 | 0.32 |
ENST00000564628.1
ENST00000563498.1 |
NME3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr1_+_154975110 | 0.31 |
ENST00000535420.1
ENST00000368426.3 |
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chrM_+_12331 | 0.29 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr5_+_67511524 | 0.28 |
ENST00000521381.1
ENST00000521657.1 |
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr12_-_101801505 | 0.27 |
ENST00000539055.1
ENST00000551688.1 ENST00000551671.1 ENST00000261636.8 |
ARL1
|
ADP-ribosylation factor-like 1 |
| chr1_-_155658260 | 0.27 |
ENST00000368339.5
ENST00000405763.3 ENST00000368340.5 ENST00000454523.1 ENST00000443231.1 ENST00000347088.5 ENST00000361831.5 ENST00000355499.4 |
YY1AP1
|
YY1 associated protein 1 |
| chr14_-_102704783 | 0.26 |
ENST00000522534.1
|
MOK
|
MOK protein kinase |
| chrX_+_47077632 | 0.26 |
ENST00000457458.2
|
CDK16
|
cyclin-dependent kinase 16 |
| chr16_-_1821721 | 0.25 |
ENST00000219302.3
|
NME3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr19_-_12267524 | 0.25 |
ENST00000455799.1
ENST00000355738.1 ENST00000439556.2 ENST00000542938.1 |
ZNF625
|
zinc finger protein 625 |
| chr17_+_46970127 | 0.25 |
ENST00000355938.5
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr6_-_31869769 | 0.25 |
ENST00000375527.2
|
ZBTB12
|
zinc finger and BTB domain containing 12 |
| chr1_+_154229547 | 0.24 |
ENST00000428595.1
|
UBAP2L
|
ubiquitin associated protein 2-like |
| chr17_+_46970178 | 0.23 |
ENST00000393366.2
ENST00000506855.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr8_+_118147498 | 0.23 |
ENST00000519688.1
ENST00000456015.2 |
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8 |
| chr11_-_35440579 | 0.23 |
ENST00000606205.1
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr12_+_66217911 | 0.23 |
ENST00000403681.2
|
HMGA2
|
high mobility group AT-hook 2 |
| chr9_+_4662282 | 0.23 |
ENST00000381883.2
|
PPAPDC2
|
phosphatidic acid phosphatase type 2 domain containing 2 |
| chr12_-_53614155 | 0.22 |
ENST00000543726.1
|
RARG
|
retinoic acid receptor, gamma |
| chr17_-_42143963 | 0.22 |
ENST00000585388.1
ENST00000293406.3 |
LSM12
|
LSM12 homolog (S. cerevisiae) |
| chr12_-_53614043 | 0.21 |
ENST00000338561.5
|
RARG
|
retinoic acid receptor, gamma |
| chr3_+_130613226 | 0.20 |
ENST00000509662.1
ENST00000328560.8 ENST00000428331.2 ENST00000359644.3 ENST00000422190.2 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr10_-_103603568 | 0.20 |
ENST00000356640.2
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr16_-_58004992 | 0.19 |
ENST00000564448.1
ENST00000251102.8 ENST00000311183.4 |
CNGB1
|
cyclic nucleotide gated channel beta 1 |
| chr17_+_46970134 | 0.17 |
ENST00000503641.1
ENST00000514808.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr1_-_155658518 | 0.17 |
ENST00000404643.1
ENST00000359205.5 ENST00000407221.1 |
YY1AP1
|
YY1 associated protein 1 |
| chr3_+_51422478 | 0.17 |
ENST00000528157.1
|
MANF
|
mesencephalic astrocyte-derived neurotrophic factor |
| chrX_-_139587225 | 0.16 |
ENST00000370536.2
|
SOX3
|
SRY (sex determining region Y)-box 3 |
| chr3_-_195538728 | 0.16 |
ENST00000349607.4
ENST00000346145.4 |
MUC4
|
mucin 4, cell surface associated |
| chr12_+_54955235 | 0.16 |
ENST00000550620.1
|
PDE1B
|
phosphodiesterase 1B, calmodulin-dependent |
| chr18_-_33077556 | 0.16 |
ENST00000589273.1
ENST00000586489.1 |
INO80C
|
INO80 complex subunit C |
| chr2_-_220173685 | 0.15 |
ENST00000423636.2
ENST00000442029.1 ENST00000412847.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr1_-_235491462 | 0.15 |
ENST00000418304.1
ENST00000264183.3 ENST00000349213.3 |
ARID4B
|
AT rich interactive domain 4B (RBP1-like) |
| chr7_-_102252038 | 0.15 |
ENST00000461209.1
|
RASA4
|
RAS p21 protein activator 4 |
| chr13_-_54706954 | 0.15 |
ENST00000606706.1
ENST00000607494.1 ENST00000427299.2 ENST00000423442.2 ENST00000451744.1 |
LINC00458
|
long intergenic non-protein coding RNA 458 |
| chr15_-_75017711 | 0.15 |
ENST00000567032.1
ENST00000564596.1 ENST00000566503.1 ENST00000395049.4 ENST00000395048.2 ENST00000379727.3 |
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1 |
| chr1_-_155658766 | 0.15 |
ENST00000295566.4
ENST00000368330.2 |
YY1AP1
|
YY1 associated protein 1 |
| chr1_+_155658849 | 0.15 |
ENST00000368336.5
ENST00000343043.3 ENST00000421487.2 ENST00000535183.1 ENST00000465375.1 ENST00000470830.1 |
DAP3
|
death associated protein 3 |
| chr9_-_112970436 | 0.14 |
ENST00000400613.4
|
C9orf152
|
chromosome 9 open reading frame 152 |
| chr2_+_219283815 | 0.14 |
ENST00000248444.5
ENST00000454069.1 ENST00000392114.2 |
VIL1
|
villin 1 |
| chr6_-_31509714 | 0.14 |
ENST00000456662.1
ENST00000431908.1 ENST00000456976.1 ENST00000428450.1 ENST00000453105.2 ENST00000418897.1 ENST00000415382.2 ENST00000449074.2 ENST00000419020.1 ENST00000428098.1 |
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr1_-_155658085 | 0.13 |
ENST00000311573.5
ENST00000438245.2 |
YY1AP1
|
YY1 associated protein 1 |
| chr12_-_6798410 | 0.13 |
ENST00000361959.3
ENST00000436774.2 ENST00000544482.1 |
ZNF384
|
zinc finger protein 384 |
| chrX_+_103411189 | 0.13 |
ENST00000493442.1
|
FAM199X
|
family with sequence similarity 199, X-linked |
| chr18_+_32558208 | 0.13 |
ENST00000436190.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr1_+_145575980 | 0.13 |
ENST00000393045.2
|
PIAS3
|
protein inhibitor of activated STAT, 3 |
| chr7_+_130020932 | 0.13 |
ENST00000484324.1
|
CPA1
|
carboxypeptidase A1 (pancreatic) |
| chr1_+_145576007 | 0.13 |
ENST00000369298.1
|
PIAS3
|
protein inhibitor of activated STAT, 3 |
| chr12_-_6798523 | 0.13 |
ENST00000319770.3
|
ZNF384
|
zinc finger protein 384 |
| chr17_+_2496971 | 0.12 |
ENST00000397195.5
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa) |
| chr12_-_6798616 | 0.11 |
ENST00000355772.4
ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384
|
zinc finger protein 384 |
| chr12_-_52715179 | 0.10 |
ENST00000293670.3
|
KRT83
|
keratin 83 |
| chr2_-_47168850 | 0.07 |
ENST00000409207.1
|
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr17_+_6679528 | 0.07 |
ENST00000321535.4
|
FBXO39
|
F-box protein 39 |
| chr7_+_5322561 | 0.07 |
ENST00000396872.3
ENST00000444741.1 ENST00000297195.4 ENST00000406453.3 |
SLC29A4
|
solute carrier family 29 (equilibrative nucleoside transporter), member 4 |
| chr11_-_125648690 | 0.07 |
ENST00000436890.2
ENST00000358524.3 |
PATE2
|
prostate and testis expressed 2 |
| chr6_-_31510181 | 0.06 |
ENST00000458640.1
ENST00000396172.1 ENST00000417556.2 |
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr16_+_71560023 | 0.06 |
ENST00000572450.1
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
| chr3_-_195538760 | 0.05 |
ENST00000475231.1
|
MUC4
|
mucin 4, cell surface associated |
| chr20_-_6034672 | 0.04 |
ENST00000378858.4
|
LRRN4
|
leucine rich repeat neuronal 4 |
| chr15_-_49103235 | 0.04 |
ENST00000380950.2
|
CEP152
|
centrosomal protein 152kDa |
| chr14_+_21498666 | 0.03 |
ENST00000481535.1
|
TPPP2
|
tubulin polymerization-promoting protein family member 2 |
| chr15_+_41913690 | 0.03 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chrX_-_107225600 | 0.02 |
ENST00000302917.1
|
TEX13B
|
testis expressed 13B |
| chr19_-_42724261 | 0.02 |
ENST00000595337.1
|
DEDD2
|
death effector domain containing 2 |
| chr5_+_149569520 | 0.02 |
ENST00000230671.2
ENST00000524041.1 |
SLC6A7
|
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr16_-_30381580 | 0.02 |
ENST00000409939.3
|
TBC1D10B
|
TBC1 domain family, member 10B |
| chr1_+_235491714 | 0.01 |
ENST00000471812.1
ENST00000358966.2 ENST00000282841.5 ENST00000391855.2 |
GGPS1
|
geranylgeranyl diphosphate synthase 1 |
| chr14_+_21498360 | 0.01 |
ENST00000321760.6
ENST00000460647.2 ENST00000530140.2 ENST00000472458.1 |
TPPP2
|
tubulin polymerization-promoting protein family member 2 |
| chr19_-_7764281 | 0.00 |
ENST00000360067.4
|
FCER2
|
Fc fragment of IgE, low affinity II, receptor for (CD23) |
| chr1_+_206317450 | 0.00 |
ENST00000358184.2
ENST00000361052.3 ENST00000360218.2 |
CTSE
|
cathepsin E |
| chr1_-_161193349 | 0.00 |
ENST00000469730.2
ENST00000463273.1 ENST00000464492.1 ENST00000367990.3 ENST00000470459.2 ENST00000468465.1 ENST00000463812.1 |
APOA2
|
apolipoprotein A-II |
| chr2_+_17721920 | 0.00 |
ENST00000295156.4
|
VSNL1
|
visinin-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.4 | 2.5 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 0.6 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.1 | 0.7 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.3 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.1 | 0.4 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 | 0.3 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.2 | GO:0031049 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.0 | 0.1 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.6 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.2 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.2 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.5 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.7 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.7 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.2 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.0 | 0.3 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.5 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.3 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.9 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.2 | 0.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 2.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.6 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.1 | 0.2 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.1 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.2 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0046923 | A-type (transient outward) potassium channel activity(GO:0005250) ER retention sequence binding(GO:0046923) |
| 0.0 | 0.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |