Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
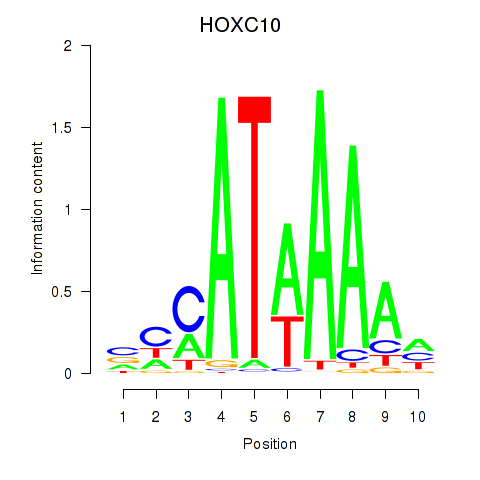
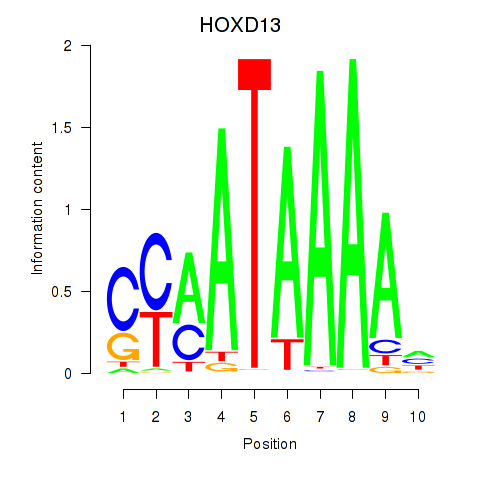
Results for HOXC10_HOXD13
Z-value: 0.77
Transcription factors associated with HOXC10_HOXD13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC10
|
ENSG00000180818.4 | homeobox C10 |
|
HOXD13
|
ENSG00000128714.5 | homeobox D13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD13 | hg19_v2_chr2_+_176957619_176957619 | -0.17 | 4.2e-01 | Click! |
| HOXC10 | hg19_v2_chr12_+_54378923_54378970 | 0.15 | 4.6e-01 | Click! |
Activity profile of HOXC10_HOXD13 motif
Sorted Z-values of HOXC10_HOXD13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_132891461 | 1.39 |
ENST00000275198.1
|
TAAR6
|
trace amine associated receptor 6 |
| chr14_-_72458326 | 1.15 |
ENST00000542853.1
|
AC005477.1
|
AC005477.1 |
| chr17_-_39646116 | 1.12 |
ENST00000328119.6
|
KRT36
|
keratin 36 |
| chr1_-_9129895 | 1.04 |
ENST00000473209.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr1_+_84630645 | 0.81 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr8_+_42873548 | 0.73 |
ENST00000533338.1
ENST00000534420.1 |
HOOK3
RP11-598P20.5
|
hook microtubule-tethering protein 3 Uncharacterized protein |
| chr9_+_82188077 | 0.72 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr2_+_109204743 | 0.72 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr17_-_46690839 | 0.72 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr8_+_76452097 | 0.69 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr20_+_58179582 | 0.67 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr8_+_38831683 | 0.60 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chr3_-_112127981 | 0.55 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr2_-_209010874 | 0.55 |
ENST00000260988.4
|
CRYGB
|
crystallin, gamma B |
| chr5_+_147582387 | 0.55 |
ENST00000325630.2
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr12_-_91576429 | 0.52 |
ENST00000552145.1
ENST00000546745.1 |
DCN
|
decorin |
| chr12_-_91573132 | 0.51 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr19_+_46367518 | 0.51 |
ENST00000302177.2
|
FOXA3
|
forkhead box A3 |
| chrX_-_19688475 | 0.50 |
ENST00000541422.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr9_+_82187630 | 0.49 |
ENST00000265284.6
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr1_-_9129631 | 0.49 |
ENST00000377414.3
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr9_+_82187487 | 0.48 |
ENST00000435650.1
ENST00000414465.1 ENST00000376537.4 ENST00000376534.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr12_-_18243119 | 0.48 |
ENST00000538724.1
ENST00000229002.2 |
RERGL
|
RERG/RAS-like |
| chr2_+_106679690 | 0.47 |
ENST00000409944.1
|
C2orf40
|
chromosome 2 open reading frame 40 |
| chr7_+_134551583 | 0.46 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr17_+_58018269 | 0.42 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr12_-_95510743 | 0.42 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr1_+_116654376 | 0.41 |
ENST00000369500.3
|
MAB21L3
|
mab-21-like 3 (C. elegans) |
| chr4_-_164534657 | 0.40 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr1_+_26485511 | 0.39 |
ENST00000374268.3
|
FAM110D
|
family with sequence similarity 110, member D |
| chrX_+_135388147 | 0.38 |
ENST00000394141.1
|
GPR112
|
G protein-coupled receptor 112 |
| chr3_-_185270342 | 0.38 |
ENST00000424591.2
|
LIPH
|
lipase, member H |
| chr6_+_12007897 | 0.38 |
ENST00000437559.1
|
RP11-456H18.2
|
RP11-456H18.2 |
| chr11_-_7961141 | 0.37 |
ENST00000360759.3
|
OR10A3
|
olfactory receptor, family 10, subfamily A, member 3 |
| chr12_+_130646999 | 0.37 |
ENST00000539839.1
ENST00000229030.4 |
FZD10
|
frizzled family receptor 10 |
| chr1_+_84630053 | 0.37 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr1_+_152635854 | 0.37 |
ENST00000368784.1
|
LCE2D
|
late cornified envelope 2D |
| chr12_-_58220078 | 0.36 |
ENST00000549039.1
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chrX_+_65382433 | 0.36 |
ENST00000374727.3
|
HEPH
|
hephaestin |
| chr1_-_76398077 | 0.36 |
ENST00000284142.6
|
ASB17
|
ankyrin repeat and SOCS box containing 17 |
| chr1_-_9129598 | 0.36 |
ENST00000535586.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr1_+_12976450 | 0.35 |
ENST00000361079.2
|
PRAMEF7
|
PRAME family member 7 |
| chr5_-_111093759 | 0.35 |
ENST00000509979.1
ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP
|
neuronal regeneration related protein |
| chr9_+_21967137 | 0.35 |
ENST00000441769.2
|
C9orf53
|
chromosome 9 open reading frame 53 |
| chrX_+_65382381 | 0.35 |
ENST00000519389.1
|
HEPH
|
hephaestin |
| chr12_-_81992111 | 0.35 |
ENST00000443686.3
ENST00000407050.4 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr3_+_68053359 | 0.34 |
ENST00000478136.1
|
FAM19A1
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 |
| chr12_+_20963647 | 0.34 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr12_+_20963632 | 0.32 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr22_+_41956767 | 0.32 |
ENST00000306149.7
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr18_-_12656715 | 0.30 |
ENST00000462226.1
ENST00000497844.2 ENST00000309836.5 ENST00000453447.2 |
SPIRE1
|
spire-type actin nucleation factor 1 |
| chr10_+_115614370 | 0.29 |
ENST00000369301.3
|
NHLRC2
|
NHL repeat containing 2 |
| chr14_+_31046959 | 0.29 |
ENST00000547532.1
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr10_+_60759378 | 0.29 |
ENST00000432535.1
|
LINC00844
|
long intergenic non-protein coding RNA 844 |
| chr1_+_209859510 | 0.28 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr18_-_52989217 | 0.27 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr7_+_80275752 | 0.27 |
ENST00000419819.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr2_-_216300784 | 0.27 |
ENST00000421182.1
ENST00000432072.2 ENST00000323926.6 ENST00000336916.4 ENST00000357867.4 ENST00000359671.1 ENST00000346544.3 ENST00000345488.5 ENST00000357009.2 ENST00000446046.1 ENST00000356005.4 ENST00000443816.1 ENST00000426059.1 ENST00000354785.4 |
FN1
|
fibronectin 1 |
| chr2_+_109237717 | 0.27 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr11_-_19082216 | 0.27 |
ENST00000329773.2
|
MRGPRX2
|
MAS-related GPR, member X2 |
| chr14_-_51027838 | 0.27 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr6_-_27775694 | 0.26 |
ENST00000377401.2
|
HIST1H2BL
|
histone cluster 1, H2bl |
| chr9_+_79074068 | 0.26 |
ENST00000444201.2
ENST00000376730.4 |
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr18_+_61223393 | 0.26 |
ENST00000269491.1
ENST00000382768.1 |
SERPINB12
|
serpin peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr17_-_53809473 | 0.26 |
ENST00000575734.1
|
TMEM100
|
transmembrane protein 100 |
| chr12_-_49418407 | 0.26 |
ENST00000526209.1
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr7_+_80275621 | 0.26 |
ENST00000426978.1
ENST00000432207.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chrX_-_117119243 | 0.25 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr3_+_15045419 | 0.25 |
ENST00000406272.2
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr17_+_35851570 | 0.25 |
ENST00000394386.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr12_+_59989918 | 0.25 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr3_+_57875711 | 0.25 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chrX_-_30327495 | 0.25 |
ENST00000453287.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr20_+_42984330 | 0.25 |
ENST00000316673.4
ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr7_+_80275953 | 0.24 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr9_-_114521783 | 0.24 |
ENST00000394779.3
ENST00000394777.4 |
C9orf84
|
chromosome 9 open reading frame 84 |
| chr11_-_47447970 | 0.24 |
ENST00000298852.3
ENST00000530912.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr12_-_15038779 | 0.24 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr11_-_47447767 | 0.24 |
ENST00000530651.1
ENST00000524447.2 ENST00000531051.2 ENST00000526993.1 ENST00000602866.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr7_-_142139783 | 0.23 |
ENST00000390374.3
|
TRBV7-6
|
T cell receptor beta variable 7-6 |
| chr20_+_52105495 | 0.23 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr3_-_141747950 | 0.23 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr5_-_111093167 | 0.23 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr2_+_242289502 | 0.23 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chr10_-_50970322 | 0.23 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr11_-_5537920 | 0.23 |
ENST00000380184.1
|
UBQLNL
|
ubiquilin-like |
| chr18_-_53069419 | 0.23 |
ENST00000570177.2
|
TCF4
|
transcription factor 4 |
| chr11_+_101785727 | 0.22 |
ENST00000263468.8
|
KIAA1377
|
KIAA1377 |
| chr12_-_58027138 | 0.22 |
ENST00000341156.4
|
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr20_+_37554955 | 0.22 |
ENST00000217429.4
|
FAM83D
|
family with sequence similarity 83, member D |
| chr1_-_167059830 | 0.22 |
ENST00000367868.3
|
GPA33
|
glycoprotein A33 (transmembrane) |
| chr6_-_137815524 | 0.22 |
ENST00000367734.2
|
OLIG3
|
oligodendrocyte transcription factor 3 |
| chr10_-_116444371 | 0.22 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr7_-_11871815 | 0.22 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chr3_+_57881966 | 0.22 |
ENST00000495364.1
|
SLMAP
|
sarcolemma associated protein |
| chr16_+_333152 | 0.22 |
ENST00000219406.6
ENST00000404312.1 ENST00000456379.1 |
PDIA2
|
protein disulfide isomerase family A, member 2 |
| chr6_+_155470243 | 0.22 |
ENST00000456877.2
ENST00000528391.2 |
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr2_+_90198535 | 0.21 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr22_+_35462129 | 0.21 |
ENST00000404699.2
ENST00000308700.6 |
ISX
|
intestine-specific homeobox |
| chr9_-_21202204 | 0.21 |
ENST00000239347.3
|
IFNA7
|
interferon, alpha 7 |
| chr7_+_108210012 | 0.21 |
ENST00000249356.3
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr2_-_165424973 | 0.21 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr16_+_25228242 | 0.21 |
ENST00000219660.5
|
AQP8
|
aquaporin 8 |
| chr6_+_108487245 | 0.20 |
ENST00000368986.4
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr12_-_81763127 | 0.20 |
ENST00000541017.1
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr12_-_81763184 | 0.20 |
ENST00000548670.1
ENST00000541570.2 ENST00000553058.1 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr1_-_161207875 | 0.20 |
ENST00000512372.1
ENST00000437437.2 ENST00000442691.2 ENST00000412844.2 ENST00000428574.2 ENST00000505005.1 ENST00000508740.1 ENST00000508387.1 ENST00000504010.1 ENST00000511676.1 ENST00000502985.1 ENST00000367981.3 ENST00000515621.1 ENST00000511944.1 ENST00000511748.1 ENST00000367984.4 ENST00000367985.3 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr19_+_36706024 | 0.20 |
ENST00000443387.2
|
ZNF146
|
zinc finger protein 146 |
| chr5_-_111312622 | 0.20 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr1_+_75600567 | 0.20 |
ENST00000356261.3
|
LHX8
|
LIM homeobox 8 |
| chr10_+_21823079 | 0.20 |
ENST00000377100.3
ENST00000377072.3 ENST00000446906.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr1_-_227505289 | 0.20 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr12_+_52695617 | 0.19 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr2_+_179149636 | 0.19 |
ENST00000409631.1
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr3_-_151034734 | 0.19 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chrX_+_16668278 | 0.19 |
ENST00000380200.3
|
S100G
|
S100 calcium binding protein G |
| chr21_-_39493433 | 0.19 |
ENST00000398948.1
ENST00000328264.3 |
DSCR4
|
Down syndrome critical region gene 4 |
| chr1_-_161207953 | 0.19 |
ENST00000367982.4
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr6_-_116833500 | 0.19 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr21_-_31859755 | 0.19 |
ENST00000334055.3
|
KRTAP19-2
|
keratin associated protein 19-2 |
| chr5_-_43412418 | 0.19 |
ENST00000537013.1
ENST00000361115.4 |
CCL28
|
chemokine (C-C motif) ligand 28 |
| chr1_+_211432775 | 0.18 |
ENST00000419091.2
|
RCOR3
|
REST corepressor 3 |
| chr19_-_19626838 | 0.18 |
ENST00000360913.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr21_+_35736302 | 0.18 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chr11_-_76155700 | 0.18 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr6_-_26247259 | 0.18 |
ENST00000244537.4
|
HIST1H4G
|
histone cluster 1, H4g |
| chr9_-_124990680 | 0.18 |
ENST00000541397.2
ENST00000560485.1 |
LHX6
|
LIM homeobox 6 |
| chr7_+_77428066 | 0.18 |
ENST00000422959.2
ENST00000307305.8 ENST00000424760.1 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr17_-_34417479 | 0.17 |
ENST00000225245.5
|
CCL3
|
chemokine (C-C motif) ligand 3 |
| chr22_+_39916558 | 0.17 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr7_+_1727755 | 0.17 |
ENST00000424383.2
|
ELFN1
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1 |
| chr3_-_123710199 | 0.17 |
ENST00000184183.4
|
ROPN1
|
rhophilin associated tail protein 1 |
| chr19_-_49843539 | 0.17 |
ENST00000602554.1
ENST00000358234.4 |
CTC-301O7.4
|
CTC-301O7.4 |
| chr1_+_32042105 | 0.17 |
ENST00000457433.2
ENST00000441210.2 |
TINAGL1
|
tubulointerstitial nephritis antigen-like 1 |
| chr12_+_48099858 | 0.17 |
ENST00000547799.1
|
RP1-197B17.3
|
RP1-197B17.3 |
| chr10_-_50970382 | 0.17 |
ENST00000419399.1
ENST00000432695.1 |
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr20_+_30193083 | 0.16 |
ENST00000376112.3
ENST00000376105.3 |
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein |
| chr8_+_52730143 | 0.16 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr11_+_64879317 | 0.16 |
ENST00000526809.1
ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2
|
transmembrane 7 superfamily member 2 |
| chr1_+_32042131 | 0.16 |
ENST00000271064.7
ENST00000537531.1 |
TINAGL1
|
tubulointerstitial nephritis antigen-like 1 |
| chr16_-_20338748 | 0.16 |
ENST00000575582.1
ENST00000341642.5 ENST00000381362.4 ENST00000572347.1 ENST00000572478.1 ENST00000302555.5 |
GP2
|
glycoprotein 2 (zymogen granule membrane) |
| chr1_-_161208013 | 0.16 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr4_-_39033963 | 0.16 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr7_-_72992865 | 0.16 |
ENST00000452475.1
|
TBL2
|
transducin (beta)-like 2 |
| chr16_-_20339123 | 0.16 |
ENST00000381360.5
|
GP2
|
glycoprotein 2 (zymogen granule membrane) |
| chr9_+_44868935 | 0.16 |
ENST00000448436.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr17_+_34391625 | 0.16 |
ENST00000004921.3
|
CCL18
|
chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated) |
| chr1_+_211432593 | 0.16 |
ENST00000367006.4
|
RCOR3
|
REST corepressor 3 |
| chr5_-_94417314 | 0.16 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr5_+_7654057 | 0.16 |
ENST00000537121.1
|
ADCY2
|
adenylate cyclase 2 (brain) |
| chrX_+_65384182 | 0.16 |
ENST00000441993.2
ENST00000419594.1 |
HEPH
|
hephaestin |
| chr14_+_23012122 | 0.15 |
ENST00000390534.1
|
TRAJ3
|
T cell receptor alpha joining 3 |
| chr1_-_161207986 | 0.15 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr7_+_134430212 | 0.15 |
ENST00000436461.2
|
CALD1
|
caldesmon 1 |
| chr17_-_38821373 | 0.15 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr2_+_169923577 | 0.15 |
ENST00000432060.2
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr20_+_3052264 | 0.15 |
ENST00000217386.2
|
OXT
|
oxytocin/neurophysin I prepropeptide |
| chrX_+_65384052 | 0.15 |
ENST00000336279.5
ENST00000458621.1 |
HEPH
|
hephaestin |
| chr12_-_96184533 | 0.15 |
ENST00000343702.4
ENST00000344911.4 |
NTN4
|
netrin 4 |
| chr15_+_25101698 | 0.15 |
ENST00000400097.1
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chr3_-_12200851 | 0.15 |
ENST00000287814.4
|
TIMP4
|
TIMP metallopeptidase inhibitor 4 |
| chrX_-_108725301 | 0.15 |
ENST00000218006.2
|
GUCY2F
|
guanylate cyclase 2F, retinal |
| chr3_+_107096188 | 0.15 |
ENST00000261058.1
|
CCDC54
|
coiled-coil domain containing 54 |
| chr11_+_118401706 | 0.14 |
ENST00000411589.2
ENST00000442938.2 ENST00000359862.4 |
TMEM25
|
transmembrane protein 25 |
| chr7_-_36634181 | 0.14 |
ENST00000538464.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr1_+_52082751 | 0.14 |
ENST00000447887.1
ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9
|
oxysterol binding protein-like 9 |
| chr19_-_54676884 | 0.14 |
ENST00000376591.4
|
TMC4
|
transmembrane channel-like 4 |
| chr5_+_147582348 | 0.14 |
ENST00000514389.1
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr19_-_54676846 | 0.14 |
ENST00000301187.4
|
TMC4
|
transmembrane channel-like 4 |
| chr12_-_91576561 | 0.14 |
ENST00000547568.2
ENST00000552962.1 |
DCN
|
decorin |
| chr3_+_57882024 | 0.14 |
ENST00000494088.1
|
SLMAP
|
sarcolemma associated protein |
| chr21_-_31588365 | 0.14 |
ENST00000399899.1
|
CLDN8
|
claudin 8 |
| chr12_-_117799446 | 0.14 |
ENST00000317775.6
ENST00000344089.3 |
NOS1
|
nitric oxide synthase 1 (neuronal) |
| chr19_-_10420459 | 0.14 |
ENST00000403352.1
ENST00000403903.3 |
ZGLP1
|
zinc finger, GATA-like protein 1 |
| chr11_+_118401899 | 0.14 |
ENST00000528373.1
ENST00000544878.1 ENST00000354284.4 ENST00000533137.1 ENST00000532762.1 ENST00000526973.1 ENST00000354064.7 ENST00000533102.1 ENST00000313236.5 ENST00000527267.1 ENST00000524725.1 ENST00000533689.1 |
TMEM25
|
transmembrane protein 25 |
| chr3_-_49314640 | 0.14 |
ENST00000436325.1
|
C3orf62
|
chromosome 3 open reading frame 62 |
| chr11_-_76155618 | 0.14 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr19_-_19626351 | 0.14 |
ENST00000585580.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chrX_+_84498989 | 0.14 |
ENST00000395402.1
|
ZNF711
|
zinc finger protein 711 |
| chr4_-_110723134 | 0.14 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr4_-_110723194 | 0.14 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr3_+_30647994 | 0.13 |
ENST00000295754.5
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr12_+_93096759 | 0.13 |
ENST00000544406.2
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr12_+_52056548 | 0.13 |
ENST00000545061.1
ENST00000355133.3 |
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr2_+_138722028 | 0.13 |
ENST00000280096.5
|
HNMT
|
histamine N-methyltransferase |
| chr12_-_111358372 | 0.13 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr17_-_39471947 | 0.13 |
ENST00000334202.3
|
KRTAP17-1
|
keratin associated protein 17-1 |
| chr15_-_58571445 | 0.13 |
ENST00000558231.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chrX_+_84499038 | 0.13 |
ENST00000373165.3
|
ZNF711
|
zinc finger protein 711 |
| chr4_+_169013666 | 0.13 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr5_-_107703556 | 0.13 |
ENST00000496714.1
|
FBXL17
|
F-box and leucine-rich repeat protein 17 |
| chr21_+_34398153 | 0.13 |
ENST00000382357.3
ENST00000430860.1 ENST00000333337.3 |
OLIG2
|
oligodendrocyte lineage transcription factor 2 |
| chr4_-_110723335 | 0.13 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr10_-_69597828 | 0.13 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr14_-_106926724 | 0.13 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr17_-_39553844 | 0.13 |
ENST00000251645.2
|
KRT31
|
keratin 31 |
| chr20_+_16729003 | 0.13 |
ENST00000246081.2
|
OTOR
|
otoraplin |
| chr9_+_75229616 | 0.13 |
ENST00000340019.3
|
TMC1
|
transmembrane channel-like 1 |
| chr6_+_88054530 | 0.13 |
ENST00000388923.4
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chr14_-_55369525 | 0.13 |
ENST00000543643.2
ENST00000536224.2 ENST00000395514.1 ENST00000491895.2 |
GCH1
|
GTP cyclohydrolase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC10_HOXD13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0097319 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.2 | 1.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.8 | GO:2000332 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.7 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.3 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.1 | 1.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.2 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.1 | 0.2 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.1 | 0.5 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.2 | GO:1990737 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.1 | 0.3 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.1 | 0.2 | GO:0002125 | maternal aggressive behavior(GO:0002125) positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.4 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.2 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.2 | GO:1902159 | regulation of inward rectifier potassium channel activity(GO:1901979) regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.0 | 0.2 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.3 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.2 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.7 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.3 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.0 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.0 | 0.1 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.0 | 0.3 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.2 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.7 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.3 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.0 | 1.7 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.0 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.3 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.0 | 0.4 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.3 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.3 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.8 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 | 0.1 | GO:0002086 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.2 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.0 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.4 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.3 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.0 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.1 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 0.1 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.9 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.0 | GO:1903991 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 1.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 1.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.5 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.2 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.2 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 1.8 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.0 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.3 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.2 | 1.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.7 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 1.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.3 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.3 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 0.2 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.1 | 1.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.3 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.2 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.4 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.2 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 0.7 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.2 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.2 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 2.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.2 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 1.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.2 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.0 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.0 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 1.3 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |