Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
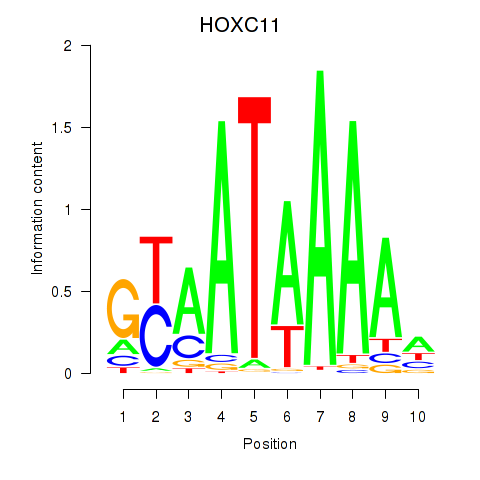
Results for HOXC11
Z-value: 0.58
Transcription factors associated with HOXC11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC11
|
ENSG00000123388.4 | homeobox C11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC11 | hg19_v2_chr12_+_54366894_54366922 | 0.22 | 2.8e-01 | Click! |
Activity profile of HOXC11 motif
Sorted Z-values of HOXC11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_32812568 | 1.58 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr7_-_47579188 | 0.91 |
ENST00000398879.1
ENST00000355730.3 ENST00000442536.2 ENST00000458317.2 |
TNS3
|
tensin 3 |
| chr3_-_79816965 | 0.90 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr6_+_127898312 | 0.84 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr7_+_35756092 | 0.82 |
ENST00000458087.3
|
AC018647.3
|
AC018647.3 |
| chr7_+_35756186 | 0.81 |
ENST00000430518.1
|
AC018647.3
|
AC018647.3 |
| chr15_+_67418047 | 0.73 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr20_+_52105495 | 0.68 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr2_+_228678550 | 0.65 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr17_+_7461613 | 0.65 |
ENST00000438470.1
ENST00000436057.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr7_-_81399411 | 0.58 |
ENST00000423064.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr3_-_37216055 | 0.57 |
ENST00000336686.4
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr12_-_52779433 | 0.57 |
ENST00000257951.3
|
KRT84
|
keratin 84 |
| chr2_-_136678123 | 0.56 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr4_-_174451370 | 0.55 |
ENST00000359562.4
|
HAND2
|
heart and neural crest derivatives expressed 2 |
| chr17_-_39646116 | 0.54 |
ENST00000328119.6
|
KRT36
|
keratin 36 |
| chr1_-_12677714 | 0.53 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr19_+_46367518 | 0.53 |
ENST00000302177.2
|
FOXA3
|
forkhead box A3 |
| chr2_-_190044480 | 0.53 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr16_+_57653989 | 0.51 |
ENST00000567835.1
ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr10_-_24770632 | 0.51 |
ENST00000596413.1
|
AL353583.1
|
AL353583.1 |
| chr16_+_57653854 | 0.50 |
ENST00000568908.1
ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr5_-_36301984 | 0.49 |
ENST00000502994.1
ENST00000515759.1 ENST00000296604.3 |
RANBP3L
|
RAN binding protein 3-like |
| chr1_-_237167718 | 0.46 |
ENST00000464121.2
|
MT1HL1
|
metallothionein 1H-like 1 |
| chr19_-_10420459 | 0.45 |
ENST00000403352.1
ENST00000403903.3 |
ZGLP1
|
zinc finger, GATA-like protein 1 |
| chr4_+_95972822 | 0.44 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr22_+_41697520 | 0.44 |
ENST00000352645.4
|
ZC3H7B
|
zinc finger CCCH-type containing 7B |
| chr2_-_145278475 | 0.42 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr1_+_209878182 | 0.41 |
ENST00000367027.3
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr7_+_108210012 | 0.37 |
ENST00000249356.3
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr17_-_77924627 | 0.36 |
ENST00000572862.1
ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16
|
TBC1 domain family, member 16 |
| chr8_+_38831683 | 0.35 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chr2_+_166428839 | 0.34 |
ENST00000342316.4
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr9_-_133814455 | 0.34 |
ENST00000448616.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr8_+_104831472 | 0.33 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr6_+_108487245 | 0.33 |
ENST00000368986.4
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr1_+_209859510 | 0.33 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr3_-_57326704 | 0.33 |
ENST00000487349.1
ENST00000389601.3 |
ASB14
|
ankyrin repeat and SOCS box containing 14 |
| chr1_-_204135450 | 0.32 |
ENST00000272190.8
ENST00000367195.2 |
REN
|
renin |
| chr1_-_9129895 | 0.32 |
ENST00000473209.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr10_-_106240032 | 0.32 |
ENST00000447860.1
|
RP11-127O4.3
|
RP11-127O4.3 |
| chr4_+_146539415 | 0.30 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr4_+_169013666 | 0.30 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr5_-_76935513 | 0.29 |
ENST00000306422.3
|
OTP
|
orthopedia homeobox |
| chr12_+_70219052 | 0.29 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chr3_-_71294304 | 0.29 |
ENST00000498215.1
|
FOXP1
|
forkhead box P1 |
| chr3_+_173116225 | 0.28 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr6_+_26156551 | 0.27 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr7_-_20826504 | 0.27 |
ENST00000418710.2
ENST00000361443.4 |
SP8
|
Sp8 transcription factor |
| chr17_-_39684550 | 0.26 |
ENST00000455635.1
ENST00000361566.3 |
KRT19
|
keratin 19 |
| chr7_-_27169801 | 0.26 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr10_-_115614127 | 0.26 |
ENST00000369305.1
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr22_+_39916558 | 0.26 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr12_+_130646999 | 0.26 |
ENST00000539839.1
ENST00000229030.4 |
FZD10
|
frizzled family receptor 10 |
| chr14_+_97925151 | 0.25 |
ENST00000554862.1
ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1
|
CTD-2506J14.1 |
| chr1_+_75600567 | 0.25 |
ENST00000356261.3
|
LHX8
|
LIM homeobox 8 |
| chr11_-_132813566 | 0.24 |
ENST00000331898.7
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr11_-_5537920 | 0.22 |
ENST00000380184.1
|
UBQLNL
|
ubiquilin-like |
| chr11_+_34664014 | 0.22 |
ENST00000527935.1
|
EHF
|
ets homologous factor |
| chr2_-_68052694 | 0.22 |
ENST00000457448.1
|
AC010987.6
|
AC010987.6 |
| chr19_+_1440838 | 0.22 |
ENST00000594262.1
|
AC027307.3
|
Uncharacterized protein |
| chr1_+_167298281 | 0.21 |
ENST00000367862.5
|
POU2F1
|
POU class 2 homeobox 1 |
| chr6_+_132873832 | 0.21 |
ENST00000275200.1
|
TAAR8
|
trace amine associated receptor 8 |
| chr11_-_118305921 | 0.20 |
ENST00000532619.1
|
RP11-770J1.4
|
RP11-770J1.4 |
| chr20_+_58713514 | 0.20 |
ENST00000432910.1
|
RP5-1043L13.1
|
RP5-1043L13.1 |
| chr6_-_64029879 | 0.20 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr1_-_152131703 | 0.20 |
ENST00000316073.3
|
RPTN
|
repetin |
| chr9_+_44868935 | 0.20 |
ENST00000448436.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr6_+_46761118 | 0.19 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr8_+_104831554 | 0.19 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr10_+_114710516 | 0.19 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr14_-_57272366 | 0.18 |
ENST00000554788.1
ENST00000554845.1 ENST00000408990.3 |
OTX2
|
orthodenticle homeobox 2 |
| chr7_-_108209897 | 0.18 |
ENST00000313516.5
|
THAP5
|
THAP domain containing 5 |
| chr12_+_25205666 | 0.18 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr14_-_101295407 | 0.17 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr12_-_111358372 | 0.17 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr8_-_18666360 | 0.17 |
ENST00000286485.8
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr20_+_58179582 | 0.17 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr3_+_63428982 | 0.17 |
ENST00000479198.1
ENST00000460711.1 ENST00000465156.1 |
SYNPR
|
synaptoporin |
| chr4_-_107957454 | 0.16 |
ENST00000285311.3
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr10_+_26505179 | 0.16 |
ENST00000376261.3
|
GAD2
|
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) |
| chr7_-_81399329 | 0.16 |
ENST00000453411.1
ENST00000444829.2 |
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr9_-_16705069 | 0.16 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr3_-_45838011 | 0.15 |
ENST00000358525.4
ENST00000413781.1 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr5_+_145718587 | 0.15 |
ENST00000230732.4
|
POU4F3
|
POU class 4 homeobox 3 |
| chr5_-_115872142 | 0.15 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr3_-_45837959 | 0.14 |
ENST00000353278.4
ENST00000456124.2 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr6_-_137815524 | 0.14 |
ENST00000367734.2
|
OLIG3
|
oligodendrocyte transcription factor 3 |
| chr5_+_176731572 | 0.14 |
ENST00000503853.1
|
PRELID1
|
PRELI domain containing 1 |
| chr14_-_57277178 | 0.14 |
ENST00000339475.5
ENST00000554559.1 ENST00000555804.1 |
OTX2
|
orthodenticle homeobox 2 |
| chr8_+_134203303 | 0.14 |
ENST00000519433.1
ENST00000517423.1 ENST00000377863.2 ENST00000220856.6 |
WISP1
|
WNT1 inducible signaling pathway protein 1 |
| chr7_+_114055052 | 0.14 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr10_-_96829246 | 0.14 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr16_-_67978016 | 0.14 |
ENST00000264005.5
|
LCAT
|
lecithin-cholesterol acyltransferase |
| chr13_-_46679185 | 0.13 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr17_-_79876010 | 0.13 |
ENST00000328666.6
|
SIRT7
|
sirtuin 7 |
| chrX_+_65382433 | 0.13 |
ENST00000374727.3
|
HEPH
|
hephaestin |
| chr14_-_25479811 | 0.13 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr13_-_46679144 | 0.13 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr12_-_47219733 | 0.13 |
ENST00000547477.1
ENST00000447411.1 ENST00000266579.4 |
SLC38A4
|
solute carrier family 38, member 4 |
| chr5_+_67586465 | 0.12 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr8_-_109799793 | 0.12 |
ENST00000297459.3
|
TMEM74
|
transmembrane protein 74 |
| chrX_+_65382381 | 0.12 |
ENST00000519389.1
|
HEPH
|
hephaestin |
| chrX_+_65384182 | 0.12 |
ENST00000441993.2
ENST00000419594.1 |
HEPH
|
hephaestin |
| chr2_-_166930131 | 0.12 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr17_+_58018269 | 0.12 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr20_+_42984330 | 0.11 |
ENST00000316673.4
ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chrX_+_65384052 | 0.11 |
ENST00000336279.5
ENST00000458621.1 |
HEPH
|
hephaestin |
| chr15_+_49715293 | 0.11 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr19_+_19626531 | 0.11 |
ENST00000507754.4
|
NDUFA13
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
| chr14_+_73706308 | 0.10 |
ENST00000554301.1
ENST00000555445.1 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr22_-_30642782 | 0.10 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr9_-_124990680 | 0.09 |
ENST00000541397.2
ENST00000560485.1 |
LHX6
|
LIM homeobox 6 |
| chr2_+_68872954 | 0.09 |
ENST00000394342.2
|
PROKR1
|
prokineticin receptor 1 |
| chrX_-_100604184 | 0.09 |
ENST00000372902.3
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast) |
| chr16_+_67906919 | 0.09 |
ENST00000358933.5
|
EDC4
|
enhancer of mRNA decapping 4 |
| chr7_-_81399355 | 0.09 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr19_-_19626838 | 0.08 |
ENST00000360913.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr8_-_114449112 | 0.08 |
ENST00000455883.2
ENST00000352409.3 ENST00000297405.5 |
CSMD3
|
CUB and Sushi multiple domains 3 |
| chr1_+_23345943 | 0.08 |
ENST00000400181.4
ENST00000542151.1 |
KDM1A
|
lysine (K)-specific demethylase 1A |
| chr7_-_81399438 | 0.08 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr21_+_35736302 | 0.08 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chr12_-_18243119 | 0.08 |
ENST00000538724.1
ENST00000229002.2 |
RERGL
|
RERG/RAS-like |
| chr5_-_34043310 | 0.08 |
ENST00000231338.7
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3 |
| chr1_+_23345930 | 0.08 |
ENST00000356634.3
|
KDM1A
|
lysine (K)-specific demethylase 1A |
| chrX_+_14547632 | 0.07 |
ENST00000218075.4
|
GLRA2
|
glycine receptor, alpha 2 |
| chr16_-_2314222 | 0.07 |
ENST00000566397.1
|
RNPS1
|
RNA binding protein S1, serine-rich domain |
| chr17_-_41739283 | 0.07 |
ENST00000393661.2
ENST00000318579.4 |
MEOX1
|
mesenchyme homeobox 1 |
| chr15_-_68497657 | 0.06 |
ENST00000448060.2
ENST00000467889.1 |
CALML4
|
calmodulin-like 4 |
| chr9_+_95736758 | 0.06 |
ENST00000337352.6
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr19_+_35634146 | 0.06 |
ENST00000586063.1
ENST00000270310.2 ENST00000588265.1 |
FXYD7
|
FXYD domain containing ion transport regulator 7 |
| chr1_-_9129735 | 0.06 |
ENST00000377424.4
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr3_-_20053741 | 0.06 |
ENST00000389050.4
|
PP2D1
|
protein phosphatase 2C-like domain containing 1 |
| chr8_-_59412717 | 0.06 |
ENST00000301645.3
|
CYP7A1
|
cytochrome P450, family 7, subfamily A, polypeptide 1 |
| chr19_-_36233332 | 0.06 |
ENST00000592537.1
ENST00000246532.1 ENST00000344990.3 ENST00000588992.1 |
IGFLR1
|
IGF-like family receptor 1 |
| chr15_+_36887069 | 0.06 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr2_+_149402553 | 0.06 |
ENST00000258484.6
ENST00000409654.1 |
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr7_-_75401513 | 0.06 |
ENST00000005180.4
|
CCL26
|
chemokine (C-C motif) ligand 26 |
| chr1_-_9129631 | 0.05 |
ENST00000377414.3
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr19_+_4402659 | 0.05 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chr12_+_52203789 | 0.05 |
ENST00000599343.1
|
AC068987.1
|
HCG1997999; cDNA FLJ33996 fis, clone DFNES2008881 |
| chr8_-_21669826 | 0.05 |
ENST00000517328.1
|
GFRA2
|
GDNF family receptor alpha 2 |
| chr11_+_34663913 | 0.05 |
ENST00000532302.1
|
EHF
|
ets homologous factor |
| chr6_-_91296602 | 0.05 |
ENST00000369325.3
ENST00000369327.3 |
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr18_+_63417864 | 0.05 |
ENST00000536984.2
|
CDH7
|
cadherin 7, type 2 |
| chr2_+_211421262 | 0.05 |
ENST00000233072.5
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr8_+_134203273 | 0.05 |
ENST00000250160.6
|
WISP1
|
WNT1 inducible signaling pathway protein 1 |
| chr12_-_24102576 | 0.05 |
ENST00000537393.1
ENST00000309359.1 ENST00000381381.2 ENST00000451604.2 |
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr1_+_183774240 | 0.04 |
ENST00000360851.3
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr6_-_91296737 | 0.04 |
ENST00000369332.3
ENST00000369329.3 |
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr5_+_148521381 | 0.04 |
ENST00000504238.1
|
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr13_-_103719196 | 0.04 |
ENST00000245312.3
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr14_-_60952739 | 0.04 |
ENST00000555476.1
ENST00000321731.3 |
C14orf39
|
chromosome 14 open reading frame 39 |
| chr19_+_36036477 | 0.04 |
ENST00000222284.5
ENST00000392204.2 |
TMEM147
|
transmembrane protein 147 |
| chr15_-_54025300 | 0.04 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr8_-_95220775 | 0.04 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr6_-_31745085 | 0.03 |
ENST00000375686.3
ENST00000447450.1 |
VWA7
|
von Willebrand factor A domain containing 7 |
| chr5_-_169626104 | 0.03 |
ENST00000520275.1
ENST00000506431.2 |
CTB-27N1.1
|
CTB-27N1.1 |
| chr17_-_46806540 | 0.03 |
ENST00000290295.7
|
HOXB13
|
homeobox B13 |
| chr16_-_21663919 | 0.03 |
ENST00000569602.1
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr7_+_134464376 | 0.03 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr2_-_207023918 | 0.03 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chrX_-_105282712 | 0.03 |
ENST00000372563.1
|
SERPINA7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr22_+_41956767 | 0.03 |
ENST00000306149.7
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr12_+_25205568 | 0.03 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr14_-_57277163 | 0.03 |
ENST00000555006.1
|
OTX2
|
orthodenticle homeobox 2 |
| chr6_-_41703952 | 0.03 |
ENST00000358871.2
ENST00000403298.4 |
TFEB
|
transcription factor EB |
| chr19_+_36036583 | 0.03 |
ENST00000392205.1
|
TMEM147
|
transmembrane protein 147 |
| chr3_+_89156799 | 0.03 |
ENST00000452448.2
ENST00000494014.1 |
EPHA3
|
EPH receptor A3 |
| chr1_-_242162375 | 0.03 |
ENST00000357246.3
|
MAP1LC3C
|
microtubule-associated protein 1 light chain 3 gamma |
| chr2_+_128293323 | 0.02 |
ENST00000389524.4
ENST00000428314.1 |
MYO7B
|
myosin VIIB |
| chr8_-_57123815 | 0.02 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr7_-_151217001 | 0.02 |
ENST00000262187.5
|
RHEB
|
Ras homolog enriched in brain |
| chr12_-_52828147 | 0.02 |
ENST00000252245.5
|
KRT75
|
keratin 75 |
| chr6_-_31745037 | 0.02 |
ENST00000375688.4
|
VWA7
|
von Willebrand factor A domain containing 7 |
| chr16_-_3350614 | 0.02 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr19_-_39924349 | 0.02 |
ENST00000602153.1
|
RPS16
|
ribosomal protein S16 |
| chr4_-_87028478 | 0.02 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr6_+_88054530 | 0.01 |
ENST00000388923.4
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chr2_+_158114051 | 0.01 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chrX_-_107682702 | 0.01 |
ENST00000372216.4
|
COL4A6
|
collagen, type IV, alpha 6 |
| chr1_+_46152886 | 0.01 |
ENST00000372025.4
|
TMEM69
|
transmembrane protein 69 |
| chr8_+_76452097 | 0.01 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr1_-_200379180 | 0.01 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr7_-_122840015 | 0.00 |
ENST00000194130.2
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporter), member 1 |
| chr1_-_7913089 | 0.00 |
ENST00000361696.5
|
UTS2
|
urotensin 2 |
| chr7_-_41742697 | 0.00 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr16_+_14805546 | 0.00 |
ENST00000552140.1
|
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr20_+_4702548 | 0.00 |
ENST00000305817.2
|
PRND
|
prion protein 2 (dublet) |
| chr13_-_81801115 | 0.00 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC11
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.2 | 0.6 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.2 | 0.6 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.2 | 0.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.7 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 1.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.3 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.5 | GO:0003219 | cardiac right ventricle formation(GO:0003219) visceral serous pericardium development(GO:0061032) |
| 0.1 | 0.3 | GO:0099545 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 | 0.4 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.3 | GO:1990737 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.1 | 0.3 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.5 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.3 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 0.3 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.1 | 0.5 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 0.3 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.4 | GO:0097319 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.1 | 0.6 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.4 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.6 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 | 0.7 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.3 | GO:0038031 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.2 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.3 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.2 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 | 0.3 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.2 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.0 | 0.1 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.1 | GO:1902159 | regulation of inward rectifier potassium channel activity(GO:1901979) regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.9 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 1.6 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) sclerotome development(GO:0061056) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.0 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.2 | 0.6 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.7 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.6 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 1.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.4 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.1 | 0.5 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.2 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.1 | 1.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.2 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.8 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.3 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 1.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |