Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for HOXD1
Z-value: 0.37
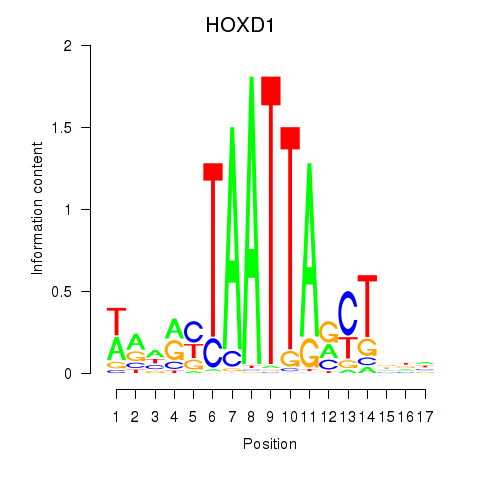
Transcription factors associated with HOXD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD1
|
ENSG00000128645.11 | homeobox D1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD1 | hg19_v2_chr2_+_177053307_177053402 | -0.09 | 6.5e-01 | Click! |
Activity profile of HOXD1 motif
Sorted Z-values of HOXD1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_102651343 | 0.64 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr4_+_144354644 | 0.53 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr4_+_86525299 | 0.49 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr5_+_66300446 | 0.47 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr20_-_50418972 | 0.41 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr20_-_50418947 | 0.40 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr15_-_55562479 | 0.32 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr15_-_55562582 | 0.31 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr15_-_55563072 | 0.29 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr4_-_119759795 | 0.28 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr3_+_111717511 | 0.27 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr3_+_111717600 | 0.26 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr3_+_111718036 | 0.24 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr1_+_117544366 | 0.23 |
ENST00000256652.4
ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr11_-_71823266 | 0.23 |
ENST00000538919.1
ENST00000539395.1 ENST00000542531.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr11_-_62521614 | 0.19 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr11_-_71823796 | 0.19 |
ENST00000545680.1
ENST00000543587.1 ENST00000538393.1 ENST00000535234.1 ENST00000227618.4 ENST00000535503.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr7_-_81399411 | 0.18 |
ENST00000423064.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr11_-_71823715 | 0.18 |
ENST00000545944.1
ENST00000502597.2 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr20_-_50419055 | 0.18 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr11_+_60163775 | 0.18 |
ENST00000300187.6
ENST00000395005.2 |
MS4A14
|
membrane-spanning 4-domains, subfamily A, member 14 |
| chr3_-_99569821 | 0.17 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr7_-_73256838 | 0.16 |
ENST00000297873.4
|
WBSCR27
|
Williams Beuren syndrome chromosome region 27 |
| chr2_+_228736335 | 0.14 |
ENST00000440997.1
ENST00000545118.1 |
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr7_-_99717463 | 0.13 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr11_-_26593779 | 0.13 |
ENST00000529533.1
|
MUC15
|
mucin 15, cell surface associated |
| chr11_-_26593677 | 0.12 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr8_+_55528627 | 0.12 |
ENST00000220676.1
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant) |
| chr20_+_43990576 | 0.12 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr13_+_36050881 | 0.11 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr18_-_33709268 | 0.11 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr11_+_71900572 | 0.11 |
ENST00000312293.4
|
FOLR1
|
folate receptor 1 (adult) |
| chr11_+_71900703 | 0.11 |
ENST00000393681.2
|
FOLR1
|
folate receptor 1 (adult) |
| chr7_+_99717230 | 0.10 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr19_+_50016610 | 0.09 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr12_+_15699286 | 0.08 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr19_-_3557570 | 0.08 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr14_+_32798462 | 0.07 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr4_-_139051839 | 0.06 |
ENST00000514600.1
ENST00000513895.1 ENST00000512536.1 |
LINC00616
|
long intergenic non-protein coding RNA 616 |
| chr8_+_92261516 | 0.06 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr4_-_74486109 | 0.06 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr7_+_133812052 | 0.06 |
ENST00000285928.2
|
LRGUK
|
leucine-rich repeats and guanylate kinase domain containing |
| chr7_-_99716952 | 0.06 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr11_+_65554493 | 0.06 |
ENST00000335987.3
|
OVOL1
|
ovo-like zinc finger 1 |
| chr4_-_74486347 | 0.05 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr5_+_176811431 | 0.05 |
ENST00000512593.1
ENST00000324417.5 |
SLC34A1
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 1 |
| chr3_-_101039402 | 0.05 |
ENST00000193391.7
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr14_+_32798547 | 0.05 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr3_+_2933893 | 0.05 |
ENST00000397459.2
|
CNTN4
|
contactin 4 |
| chr5_+_63802109 | 0.05 |
ENST00000334025.2
|
RGS7BP
|
regulator of G-protein signaling 7 binding protein |
| chr15_-_37393406 | 0.04 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr12_-_52761262 | 0.04 |
ENST00000257901.3
|
KRT85
|
keratin 85 |
| chr4_+_113568207 | 0.04 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr14_+_56584414 | 0.04 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr11_+_118398178 | 0.04 |
ENST00000302783.4
ENST00000539546.1 |
TTC36
|
tetratricopeptide repeat domain 36 |
| chr22_+_24951949 | 0.04 |
ENST00000402849.1
|
SNRPD3
|
small nuclear ribonucleoprotein D3 polypeptide 18kDa |
| chr1_-_151431909 | 0.03 |
ENST00000361398.3
ENST00000271715.2 |
POGZ
|
pogo transposable element with ZNF domain |
| chr12_-_22063787 | 0.03 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr17_-_27418537 | 0.03 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr10_+_99205894 | 0.03 |
ENST00000370854.3
ENST00000393760.1 ENST00000414567.1 ENST00000370846.4 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr4_-_25865159 | 0.02 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr1_-_190446759 | 0.02 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr3_-_151034734 | 0.02 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr3_-_164796269 | 0.02 |
ENST00000264382.3
|
SI
|
sucrase-isomaltase (alpha-glucosidase) |
| chr9_+_124329336 | 0.02 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr2_+_135596106 | 0.02 |
ENST00000356140.5
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr7_-_81399329 | 0.02 |
ENST00000453411.1
ENST00000444829.2 |
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr10_+_99205959 | 0.01 |
ENST00000352634.4
ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr12_+_109577202 | 0.01 |
ENST00000377848.3
ENST00000377854.5 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr7_-_81399438 | 0.01 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr12_-_118796910 | 0.01 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr3_-_191000172 | 0.01 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr2_+_187371440 | 0.01 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr11_+_73498898 | 0.00 |
ENST00000535529.1
ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr2_+_135596180 | 0.00 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr2_+_11682790 | 0.00 |
ENST00000389825.3
ENST00000381483.2 |
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr4_-_150736962 | 0.00 |
ENST00000502345.1
ENST00000510975.1 ENST00000511993.1 |
RP11-526A4.1
|
RP11-526A4.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.2 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.1 | 1.0 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.6 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.5 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0042670 | photoreceptor cell outer segment organization(GO:0035845) retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0071209 | U7 snRNA binding(GO:0071209) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |