Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
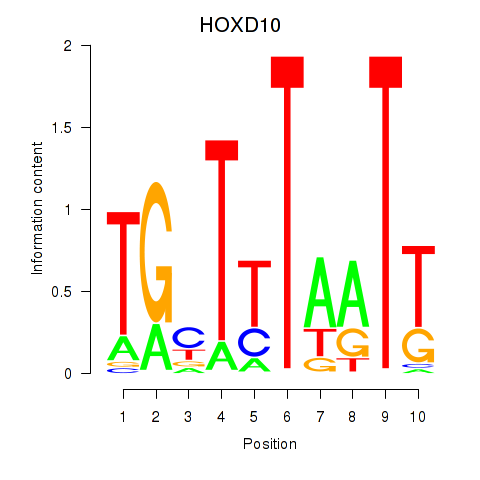
Results for HOXD10
Z-value: 0.65
Transcription factors associated with HOXD10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD10
|
ENSG00000128710.5 | homeobox D10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD10 | hg19_v2_chr2_+_176981307_176981307 | -0.62 | 8.5e-04 | Click! |
Activity profile of HOXD10 motif
Sorted Z-values of HOXD10 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_74675521 | 1.28 |
ENST00000377024.3
|
C9orf57
|
chromosome 9 open reading frame 57 |
| chr18_+_48918368 | 1.28 |
ENST00000583982.1
ENST00000578152.1 ENST00000583609.1 ENST00000435144.1 ENST00000580841.1 |
RP11-267C16.1
|
RP11-267C16.1 |
| chr6_+_147527103 | 1.18 |
ENST00000179882.6
|
STXBP5
|
syntaxin binding protein 5 (tomosyn) |
| chr18_-_22932080 | 1.04 |
ENST00000584787.1
ENST00000361524.3 ENST00000538137.2 |
ZNF521
|
zinc finger protein 521 |
| chr3_-_64211112 | 0.94 |
ENST00000295902.6
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr8_+_124084899 | 0.92 |
ENST00000287380.1
ENST00000309336.3 ENST00000519418.1 ENST00000327098.5 ENST00000522420.1 ENST00000521676.1 ENST00000378080.2 |
TBC1D31
|
TBC1 domain family, member 31 |
| chr13_+_114567131 | 0.90 |
ENST00000608651.1
|
GAS6-AS2
|
GAS6 antisense RNA 2 (head to head) |
| chr2_-_188312971 | 0.85 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr1_+_78470530 | 0.79 |
ENST00000370763.5
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr12_-_10022735 | 0.73 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr1_+_158978768 | 0.71 |
ENST00000447473.2
|
IFI16
|
interferon, gamma-inducible protein 16 |
| chr9_-_47314222 | 0.67 |
ENST00000420228.1
ENST00000438517.1 ENST00000414020.1 |
AL953854.2
|
AL953854.2 |
| chr1_-_76398077 | 0.67 |
ENST00000284142.6
|
ASB17
|
ankyrin repeat and SOCS box containing 17 |
| chr12_+_20963632 | 0.63 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr20_-_17539456 | 0.63 |
ENST00000544874.1
ENST00000377868.2 |
BFSP1
|
beaded filament structural protein 1, filensin |
| chr1_+_164528866 | 0.62 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr12_+_20963647 | 0.62 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr3_+_43328004 | 0.62 |
ENST00000454177.1
ENST00000429705.2 ENST00000296088.7 ENST00000437827.1 |
SNRK
|
SNF related kinase |
| chr2_-_165424973 | 0.61 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr4_+_86748898 | 0.61 |
ENST00000509300.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr18_-_25616519 | 0.58 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr4_-_89951028 | 0.57 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr14_-_106453155 | 0.56 |
ENST00000390594.2
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr12_-_76817036 | 0.56 |
ENST00000546946.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr5_-_55529115 | 0.55 |
ENST00000513241.2
ENST00000341048.4 |
ANKRD55
|
ankyrin repeat domain 55 |
| chr8_-_95274536 | 0.55 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr1_-_149459549 | 0.54 |
ENST00000369175.3
|
FAM72C
|
family with sequence similarity 72, member C |
| chr8_+_74903580 | 0.49 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr2_-_190627481 | 0.48 |
ENST00000264151.5
ENST00000520350.1 ENST00000521630.1 ENST00000517895.1 |
OSGEPL1
|
O-sialoglycoprotein endopeptidase-like 1 |
| chr9_+_75263565 | 0.47 |
ENST00000396237.3
|
TMC1
|
transmembrane channel-like 1 |
| chr3_+_107318157 | 0.47 |
ENST00000406780.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr21_-_35016231 | 0.47 |
ENST00000438788.1
|
CRYZL1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr2_-_89521942 | 0.46 |
ENST00000482769.1
|
IGKV2-28
|
immunoglobulin kappa variable 2-28 |
| chr1_+_62439037 | 0.46 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr2_+_47630255 | 0.44 |
ENST00000406134.1
|
MSH2
|
mutS homolog 2 |
| chr10_-_52008313 | 0.43 |
ENST00000329428.6
ENST00000395526.4 ENST00000447815.1 |
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr1_+_40713573 | 0.42 |
ENST00000372766.3
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr4_+_108910870 | 0.42 |
ENST00000403312.1
ENST00000603302.1 ENST00000309522.3 |
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr14_-_20774092 | 0.42 |
ENST00000423949.2
ENST00000553828.1 ENST00000258821.3 |
TTC5
|
tetratricopeptide repeat domain 5 |
| chr10_+_94594351 | 0.42 |
ENST00000371552.4
|
EXOC6
|
exocyst complex component 6 |
| chr6_+_96025341 | 0.42 |
ENST00000369293.1
ENST00000358812.4 |
MANEA
|
mannosidase, endo-alpha |
| chr12_-_71148413 | 0.42 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr12_-_71148357 | 0.41 |
ENST00000378778.1
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr4_+_77356248 | 0.41 |
ENST00000296043.6
|
SHROOM3
|
shroom family member 3 |
| chr5_+_131892603 | 0.40 |
ENST00000378823.3
ENST00000265335.6 |
RAD50
|
RAD50 homolog (S. cerevisiae) |
| chr13_-_67802549 | 0.40 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr4_+_86749045 | 0.38 |
ENST00000514229.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr13_+_33160553 | 0.38 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr20_+_59654146 | 0.36 |
ENST00000441660.1
|
RP5-827L5.1
|
RP5-827L5.1 |
| chr11_+_22689648 | 0.36 |
ENST00000278187.3
|
GAS2
|
growth arrest-specific 2 |
| chr4_-_90758227 | 0.36 |
ENST00000506691.1
ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr12_-_100656134 | 0.35 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chr3_+_186435065 | 0.35 |
ENST00000287611.2
ENST00000265023.4 |
KNG1
|
kininogen 1 |
| chr2_+_88047606 | 0.34 |
ENST00000359481.4
|
PLGLB2
|
plasminogen-like B2 |
| chr3_+_169629354 | 0.34 |
ENST00000428432.2
ENST00000335556.3 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr1_+_79115503 | 0.34 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chrX_-_8139308 | 0.34 |
ENST00000317103.4
|
VCX2
|
variable charge, X-linked 2 |
| chr4_+_40198527 | 0.33 |
ENST00000381799.5
|
RHOH
|
ras homolog family member H |
| chr1_-_232598163 | 0.33 |
ENST00000308942.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr1_+_78383813 | 0.33 |
ENST00000342754.5
|
NEXN
|
nexilin (F actin binding protein) |
| chr16_-_30122717 | 0.33 |
ENST00000566613.1
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr4_-_102268628 | 0.33 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr9_+_75229616 | 0.32 |
ENST00000340019.3
|
TMC1
|
transmembrane channel-like 1 |
| chr6_+_132455118 | 0.31 |
ENST00000458028.1
|
LINC01013
|
long intergenic non-protein coding RNA 1013 |
| chr15_+_57511609 | 0.31 |
ENST00000543579.1
ENST00000537840.1 ENST00000343827.3 |
TCF12
|
transcription factor 12 |
| chr4_-_102268484 | 0.31 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr12_+_79258547 | 0.31 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr16_-_71610985 | 0.30 |
ENST00000355962.4
|
TAT
|
tyrosine aminotransferase |
| chr15_+_76352178 | 0.30 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr6_-_76072719 | 0.30 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr3_-_157221128 | 0.30 |
ENST00000392833.2
ENST00000362010.2 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr2_+_89998789 | 0.30 |
ENST00000453166.2
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr6_-_8102714 | 0.29 |
ENST00000502429.1
ENST00000429723.2 ENST00000507463.1 ENST00000379715.5 |
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr2_-_211341411 | 0.29 |
ENST00000233714.4
ENST00000443314.1 ENST00000441020.3 ENST00000450366.2 ENST00000431941.2 |
LANCL1
|
LanC lantibiotic synthetase component C-like 1 (bacterial) |
| chr7_+_135611542 | 0.29 |
ENST00000416501.1
|
AC015987.2
|
AC015987.2 |
| chr11_+_7110165 | 0.29 |
ENST00000306904.5
|
RBMXL2
|
RNA binding motif protein, X-linked-like 2 |
| chr9_+_27109133 | 0.29 |
ENST00000519097.1
ENST00000380036.4 |
TEK
|
TEK tyrosine kinase, endothelial |
| chr11_+_63137251 | 0.29 |
ENST00000310969.4
ENST00000279178.3 |
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9 |
| chr11_-_77791156 | 0.28 |
ENST00000281031.4
|
NDUFC2
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa |
| chr2_+_207804278 | 0.28 |
ENST00000272852.3
|
CPO
|
carboxypeptidase O |
| chrX_+_10031499 | 0.28 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr5_+_140227048 | 0.28 |
ENST00000532602.1
|
PCDHA9
|
protocadherin alpha 9 |
| chr6_+_24667257 | 0.28 |
ENST00000537591.1
ENST00000230048.4 |
ACOT13
|
acyl-CoA thioesterase 13 |
| chr9_-_130517522 | 0.27 |
ENST00000373274.3
ENST00000420366.1 |
SH2D3C
|
SH2 domain containing 3C |
| chr11_+_120255997 | 0.27 |
ENST00000532993.1
|
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr2_-_130902567 | 0.26 |
ENST00000457413.1
ENST00000392984.3 ENST00000409128.1 ENST00000441670.1 ENST00000409943.3 ENST00000409234.3 ENST00000310463.6 |
CCDC74B
|
coiled-coil domain containing 74B |
| chr1_+_186798073 | 0.26 |
ENST00000367466.3
ENST00000442353.2 |
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr1_+_207262881 | 0.26 |
ENST00000451804.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr15_+_48498480 | 0.26 |
ENST00000380993.3
ENST00000396577.3 |
SLC12A1
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr12_-_7656357 | 0.25 |
ENST00000396620.3
ENST00000432237.2 ENST00000359156.4 |
CD163
|
CD163 molecule |
| chr12_-_118797475 | 0.25 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr6_+_121756809 | 0.25 |
ENST00000282561.3
|
GJA1
|
gap junction protein, alpha 1, 43kDa |
| chrX_+_37639302 | 0.25 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr6_-_119031228 | 0.24 |
ENST00000392500.3
ENST00000368488.5 ENST00000434604.1 |
CEP85L
|
centrosomal protein 85kDa-like |
| chr6_+_166945369 | 0.24 |
ENST00000598601.1
|
Z98049.1
|
CDNA FLJ25492 fis, clone CBR01389; Uncharacterized protein |
| chr1_-_231560790 | 0.24 |
ENST00000366641.3
|
EGLN1
|
egl-9 family hypoxia-inducible factor 1 |
| chr14_+_22771851 | 0.24 |
ENST00000390466.1
|
TRAV39
|
T cell receptor alpha variable 39 |
| chrX_+_107334895 | 0.24 |
ENST00000372232.3
ENST00000345734.3 ENST00000372254.3 |
ATG4A
|
autophagy related 4A, cysteine peptidase |
| chr1_+_15986364 | 0.24 |
ENST00000345034.1
|
RSC1A1
|
regulatory solute carrier protein, family 1, member 1 |
| chr16_-_15736881 | 0.23 |
ENST00000540441.2
|
KIAA0430
|
KIAA0430 |
| chr17_+_48823896 | 0.23 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr1_-_153113927 | 0.23 |
ENST00000368752.4
|
SPRR2B
|
small proline-rich protein 2B |
| chr5_+_140254884 | 0.23 |
ENST00000398631.2
|
PCDHA12
|
protocadherin alpha 12 |
| chr12_+_8309630 | 0.23 |
ENST00000396570.3
|
ZNF705A
|
zinc finger protein 705A |
| chr5_-_111312622 | 0.23 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr19_-_44388116 | 0.23 |
ENST00000587539.1
|
ZNF404
|
zinc finger protein 404 |
| chr5_-_36152031 | 0.22 |
ENST00000296603.4
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr7_+_77469439 | 0.22 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr10_-_120925054 | 0.22 |
ENST00000419372.1
ENST00000369131.4 ENST00000330036.6 ENST00000355697.2 |
SFXN4
|
sideroflexin 4 |
| chr15_+_41099254 | 0.22 |
ENST00000570108.1
ENST00000564258.1 ENST00000355341.4 ENST00000336455.5 |
ZFYVE19
|
zinc finger, FYVE domain containing 19 |
| chr10_+_116697946 | 0.22 |
ENST00000298746.3
|
TRUB1
|
TruB pseudouridine (psi) synthase family member 1 |
| chr18_+_3252206 | 0.22 |
ENST00000578562.2
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr12_+_118814344 | 0.22 |
ENST00000397564.2
|
SUDS3
|
suppressor of defective silencing 3 homolog (S. cerevisiae) |
| chr8_+_26150628 | 0.22 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr18_+_6729698 | 0.22 |
ENST00000383472.4
|
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr5_-_122759032 | 0.22 |
ENST00000510582.3
ENST00000328236.5 ENST00000306481.6 ENST00000508442.2 ENST00000395431.2 |
CEP120
|
centrosomal protein 120kDa |
| chr11_+_112047087 | 0.22 |
ENST00000526088.1
ENST00000532593.1 ENST00000531169.1 |
BCO2
|
beta-carotene oxygenase 2 |
| chr13_+_76378305 | 0.21 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chr12_+_118814185 | 0.21 |
ENST00000543473.1
|
SUDS3
|
suppressor of defective silencing 3 homolog (S. cerevisiae) |
| chr11_-_31531121 | 0.21 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr9_+_42671887 | 0.21 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr1_+_206972215 | 0.20 |
ENST00000340758.2
|
IL19
|
interleukin 19 |
| chr7_+_107224364 | 0.20 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr7_-_7782204 | 0.20 |
ENST00000418534.2
|
AC007161.5
|
AC007161.5 |
| chr10_+_124739964 | 0.20 |
ENST00000406217.2
|
PSTK
|
phosphoseryl-tRNA kinase |
| chrX_+_10124977 | 0.20 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr6_-_29324054 | 0.20 |
ENST00000543825.1
|
OR5V1
|
olfactory receptor, family 5, subfamily V, member 1 |
| chr12_-_10324716 | 0.20 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr14_-_82089405 | 0.20 |
ENST00000554211.1
|
RP11-799P8.1
|
RP11-799P8.1 |
| chr1_-_243326612 | 0.19 |
ENST00000492145.1
ENST00000490813.1 ENST00000464936.1 |
CEP170
|
centrosomal protein 170kDa |
| chr12_+_16109519 | 0.19 |
ENST00000526530.1
|
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr1_+_41204506 | 0.19 |
ENST00000525290.1
ENST00000530965.1 ENST00000416859.2 ENST00000308733.5 |
NFYC
|
nuclear transcription factor Y, gamma |
| chr11_-_77790865 | 0.19 |
ENST00000534029.1
ENST00000525085.1 ENST00000527806.1 ENST00000528164.1 ENST00000528251.1 ENST00000530054.1 |
NDUFC2
NDUFC2-KCTD14
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa NDUFC2-KCTD14 readthrough |
| chr10_+_695888 | 0.19 |
ENST00000441152.2
|
PRR26
|
proline rich 26 |
| chr6_-_15586238 | 0.19 |
ENST00000462989.2
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr1_+_174844645 | 0.18 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr9_+_70856397 | 0.18 |
ENST00000360171.6
|
CBWD3
|
COBW domain containing 3 |
| chr12_-_11214893 | 0.18 |
ENST00000533467.1
|
TAS2R46
|
taste receptor, type 2, member 46 |
| chr12_-_9102549 | 0.18 |
ENST00000000412.3
|
M6PR
|
mannose-6-phosphate receptor (cation dependent) |
| chr10_+_124739911 | 0.18 |
ENST00000405485.1
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr18_+_3252265 | 0.17 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr8_+_21823726 | 0.17 |
ENST00000433566.4
|
XPO7
|
exportin 7 |
| chr8_-_124279627 | 0.17 |
ENST00000357082.4
|
ZHX1-C8ORF76
|
ZHX1-C8ORF76 readthrough |
| chr4_+_130017268 | 0.17 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr2_-_87248975 | 0.17 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr5_-_135290705 | 0.17 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr10_+_35456444 | 0.16 |
ENST00000361599.4
|
CREM
|
cAMP responsive element modulator |
| chr3_+_35721182 | 0.16 |
ENST00000413378.1
ENST00000417925.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr10_+_5488564 | 0.16 |
ENST00000449083.1
ENST00000380359.3 |
NET1
|
neuroepithelial cell transforming 1 |
| chr6_-_160679905 | 0.16 |
ENST00000366953.3
|
SLC22A2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr13_+_76378357 | 0.16 |
ENST00000489941.2
ENST00000525373.1 |
LMO7
|
LIM domain 7 |
| chr5_-_111754948 | 0.16 |
ENST00000261486.5
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A |
| chr11_-_110583912 | 0.16 |
ENST00000533353.1
ENST00000527598.1 |
ARHGAP20
|
Rho GTPase activating protein 20 |
| chr6_-_26250835 | 0.16 |
ENST00000446824.2
|
HIST1H3F
|
histone cluster 1, H3f |
| chr18_-_10701979 | 0.16 |
ENST00000538948.1
ENST00000285141.4 |
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr11_-_124180733 | 0.16 |
ENST00000357821.2
|
OR8D1
|
olfactory receptor, family 8, subfamily D, member 1 |
| chr6_+_28317685 | 0.16 |
ENST00000252211.2
ENST00000341464.5 ENST00000377255.3 |
ZKSCAN3
|
zinc finger with KRAB and SCAN domains 3 |
| chr4_-_90756769 | 0.16 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr5_+_64064748 | 0.15 |
ENST00000381070.3
ENST00000508024.1 |
CWC27
|
CWC27 spliceosome-associated protein homolog (S. cerevisiae) |
| chr1_-_156399184 | 0.15 |
ENST00000368243.1
ENST00000357975.4 ENST00000310027.5 ENST00000400991.2 |
C1orf61
|
chromosome 1 open reading frame 61 |
| chr8_-_101719159 | 0.15 |
ENST00000520868.1
ENST00000522658.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr11_-_102576537 | 0.15 |
ENST00000260229.4
|
MMP27
|
matrix metallopeptidase 27 |
| chr11_-_104769141 | 0.15 |
ENST00000508062.1
ENST00000422698.2 |
CASP12
|
caspase 12 (gene/pseudogene) |
| chr1_+_248185250 | 0.15 |
ENST00000355281.1
|
OR2L5
|
olfactory receptor, family 2, subfamily L, member 5 |
| chr1_-_54405773 | 0.15 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr3_+_186435137 | 0.15 |
ENST00000447445.1
|
KNG1
|
kininogen 1 |
| chr9_-_95166841 | 0.14 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr16_+_57481382 | 0.14 |
ENST00000564655.1
ENST00000567072.1 ENST00000567933.1 ENST00000563166.1 |
COQ9
|
coenzyme Q9 |
| chr6_+_25963020 | 0.14 |
ENST00000357085.3
|
TRIM38
|
tripartite motif containing 38 |
| chr6_-_150067696 | 0.14 |
ENST00000340413.2
ENST00000367403.3 |
NUP43
|
nucleoporin 43kDa |
| chr1_+_207277590 | 0.14 |
ENST00000367070.3
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr3_-_126327398 | 0.14 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr11_+_18433840 | 0.14 |
ENST00000541669.1
ENST00000280704.4 |
LDHC
|
lactate dehydrogenase C |
| chr5_+_122181128 | 0.14 |
ENST00000261369.4
|
SNX24
|
sorting nexin 24 |
| chr12_+_60083118 | 0.14 |
ENST00000261187.4
ENST00000543448.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr3_+_158288999 | 0.14 |
ENST00000482628.1
ENST00000478894.2 ENST00000392822.3 ENST00000466246.1 |
MLF1
|
myeloid leukemia factor 1 |
| chr6_-_20212630 | 0.14 |
ENST00000324607.7
ENST00000541730.1 ENST00000536798.1 |
MBOAT1
|
membrane bound O-acyltransferase domain containing 1 |
| chr9_+_108006880 | 0.14 |
ENST00000374723.1
ENST00000374720.3 ENST00000374724.1 |
SLC44A1
|
solute carrier family 44 (choline transporter), member 1 |
| chr3_-_185538849 | 0.14 |
ENST00000421047.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr19_-_53770972 | 0.13 |
ENST00000311170.4
|
VN1R4
|
vomeronasal 1 receptor 4 |
| chr3_-_128206759 | 0.13 |
ENST00000430265.2
|
GATA2
|
GATA binding protein 2 |
| chr4_+_166300084 | 0.13 |
ENST00000402744.4
|
CPE
|
carboxypeptidase E |
| chr18_+_61445007 | 0.13 |
ENST00000447428.1
ENST00000546027.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr15_+_71389281 | 0.13 |
ENST00000355327.3
|
THSD4
|
thrombospondin, type I, domain containing 4 |
| chrX_-_41449204 | 0.13 |
ENST00000378179.3
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr1_-_242162375 | 0.13 |
ENST00000357246.3
|
MAP1LC3C
|
microtubule-associated protein 1 light chain 3 gamma |
| chr6_+_158733692 | 0.13 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr8_+_39770803 | 0.13 |
ENST00000518237.1
|
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr4_-_84035905 | 0.13 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr18_+_46065393 | 0.13 |
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr6_-_132967142 | 0.12 |
ENST00000275216.1
|
TAAR1
|
trace amine associated receptor 1 |
| chr1_-_89736434 | 0.12 |
ENST00000370459.3
|
GBP5
|
guanylate binding protein 5 |
| chr16_+_20499024 | 0.12 |
ENST00000593357.1
|
AC137056.1
|
Uncharacterized protein; cDNA FLJ34659 fis, clone KIDNE2018863 |
| chr5_+_118407053 | 0.12 |
ENST00000311085.8
ENST00000539542.1 |
DMXL1
|
Dmx-like 1 |
| chr5_-_137475071 | 0.12 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr4_+_78804393 | 0.12 |
ENST00000502384.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr1_+_244214577 | 0.12 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr18_-_19997878 | 0.12 |
ENST00000391403.2
|
CTAGE1
|
cutaneous T-cell lymphoma-associated antigen 1 |
| chr9_+_4839762 | 0.12 |
ENST00000448872.2
ENST00000441844.1 |
RCL1
|
RNA terminal phosphate cyclase-like 1 |
| chr7_+_90338712 | 0.12 |
ENST00000265741.3
ENST00000406263.1 |
CDK14
|
cyclin-dependent kinase 14 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD10
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.2 | 0.5 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.5 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.8 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.8 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.5 | GO:0051622 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.4 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.6 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 0.4 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.1 | 0.3 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 | 0.9 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 0.2 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 0.2 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.1 | 0.4 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 0.5 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 0.3 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.4 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.6 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.0 | 0.4 | GO:1903027 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 | 0.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.2 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.0 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.2 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.3 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.6 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.0 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.2 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 1.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.4 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.1 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.1 | GO:0071284 | response to platinum ion(GO:0070541) cellular response to lead ion(GO:0071284) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.2 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.1 | GO:0032252 | negative regulation of triglyceride catabolic process(GO:0010897) secretory granule localization(GO:0032252) |
| 0.0 | 0.1 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.0 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.2 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.0 | 0.1 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.0 | 0.1 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.3 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.6 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.2 | GO:0001956 | positive regulation of neurotransmitter secretion(GO:0001956) |
| 0.0 | 0.1 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.0 | 0.1 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.7 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.6 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 1.0 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.6 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.0 | 0.2 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.5 | GO:0042311 | vasodilation(GO:0042311) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.1 | 0.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.5 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.3 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.4 | GO:0000406 | heteroduplex DNA loop binding(GO:0000404) double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.1 | 0.5 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.3 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.7 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.2 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.1 | 0.5 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 1.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.5 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 0.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.7 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 1.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.4 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.8 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.4 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.1 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.9 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |