Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
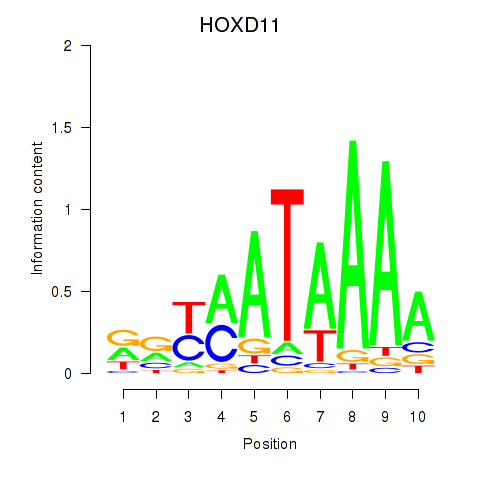
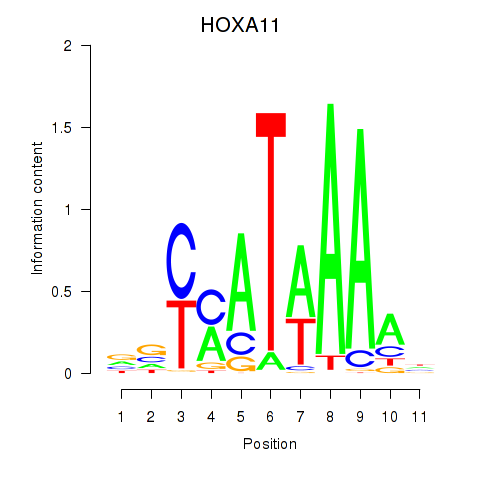
Results for HOXD11_HOXA11
Z-value: 0.72
Transcription factors associated with HOXD11_HOXA11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD11
|
ENSG00000128713.11 | homeobox D11 |
|
HOXA11
|
ENSG00000005073.5 | homeobox A11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA11 | hg19_v2_chr7_-_27224842_27224872, hg19_v2_chr7_-_27224795_27224840 | 0.22 | 2.9e-01 | Click! |
| HOXD11 | hg19_v2_chr2_+_176972000_176972025 | 0.07 | 7.4e-01 | Click! |
Activity profile of HOXD11_HOXA11 motif
Sorted Z-values of HOXD11_HOXA11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_86525299 | 1.81 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr17_-_39646116 | 1.58 |
ENST00000328119.6
|
KRT36
|
keratin 36 |
| chr6_+_32812568 | 1.57 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr21_-_31869451 | 1.30 |
ENST00000334058.2
|
KRTAP19-4
|
keratin associated protein 19-4 |
| chr3_-_79816965 | 0.87 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr4_-_76957214 | 0.82 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr10_-_24770632 | 0.80 |
ENST00000596413.1
|
AL353583.1
|
AL353583.1 |
| chr7_-_41742697 | 0.76 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr21_-_31859755 | 0.75 |
ENST00000334055.3
|
KRTAP19-2
|
keratin associated protein 19-2 |
| chr8_+_66955648 | 0.72 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr10_+_102222798 | 0.68 |
ENST00000343737.5
|
WNT8B
|
wingless-type MMTV integration site family, member 8B |
| chr11_+_34663913 | 0.62 |
ENST00000532302.1
|
EHF
|
ets homologous factor |
| chr7_+_35756092 | 0.62 |
ENST00000458087.3
|
AC018647.3
|
AC018647.3 |
| chr7_+_35756186 | 0.61 |
ENST00000430518.1
|
AC018647.3
|
AC018647.3 |
| chr11_+_120039685 | 0.60 |
ENST00000530303.1
ENST00000319763.1 |
AP000679.2
|
Uncharacterized protein |
| chr8_+_118147498 | 0.58 |
ENST00000519688.1
ENST00000456015.2 |
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8 |
| chr1_+_62439037 | 0.56 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr8_+_125985531 | 0.55 |
ENST00000319286.5
|
ZNF572
|
zinc finger protein 572 |
| chr1_+_79086088 | 0.53 |
ENST00000370751.5
ENST00000342282.3 |
IFI44L
|
interferon-induced protein 44-like |
| chr11_-_51412448 | 0.52 |
ENST00000319760.6
|
OR4A5
|
olfactory receptor, family 4, subfamily A, member 5 |
| chr10_-_115904361 | 0.52 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr17_+_42785976 | 0.50 |
ENST00000393547.2
ENST00000398338.3 |
DBF4B
|
DBF4 homolog B (S. cerevisiae) |
| chr3_-_156534754 | 0.49 |
ENST00000472943.1
ENST00000473352.1 |
LINC00886
|
long intergenic non-protein coding RNA 886 |
| chr2_+_67624430 | 0.48 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr5_-_135290705 | 0.46 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr3_+_151531810 | 0.46 |
ENST00000232892.7
|
AADAC
|
arylacetamide deacetylase |
| chr1_+_6640108 | 0.46 |
ENST00000377674.4
ENST00000488936.1 |
ZBTB48
|
zinc finger and BTB domain containing 48 |
| chr10_+_24755416 | 0.46 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr3_+_101546827 | 0.45 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr2_-_216878305 | 0.44 |
ENST00000263268.6
|
MREG
|
melanoregulin |
| chr11_+_19798964 | 0.44 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr11_-_796197 | 0.43 |
ENST00000530360.1
ENST00000528606.1 ENST00000320230.5 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr15_-_58571445 | 0.42 |
ENST00000558231.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr10_-_90712520 | 0.40 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr4_+_146539415 | 0.40 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr11_-_5173599 | 0.40 |
ENST00000328942.1
|
OR52A1
|
olfactory receptor, family 52, subfamily A, member 1 |
| chr3_-_46068969 | 0.39 |
ENST00000542109.1
ENST00000395946.2 |
XCR1
|
chemokine (C motif) receptor 1 |
| chr5_-_41261540 | 0.39 |
ENST00000263413.3
|
C6
|
complement component 6 |
| chr8_+_94241867 | 0.38 |
ENST00000598428.1
|
AC016885.1
|
Uncharacterized protein |
| chr3_+_16306837 | 0.38 |
ENST00000606098.1
|
OXNAD1
|
oxidoreductase NAD-binding domain containing 1 |
| chr19_+_46367518 | 0.38 |
ENST00000302177.2
|
FOXA3
|
forkhead box A3 |
| chr6_+_12290586 | 0.37 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr4_+_71458012 | 0.37 |
ENST00000449493.2
|
AMBN
|
ameloblastin (enamel matrix protein) |
| chr17_-_39684550 | 0.37 |
ENST00000455635.1
ENST00000361566.3 |
KRT19
|
keratin 19 |
| chr2_-_214013353 | 0.36 |
ENST00000451136.2
ENST00000421754.2 ENST00000374327.4 ENST00000413091.3 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr9_-_133814455 | 0.36 |
ENST00000448616.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr8_-_116504448 | 0.35 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr1_+_171107241 | 0.35 |
ENST00000236166.3
|
FMO6P
|
flavin containing monooxygenase 6 pseudogene |
| chrX_+_16668278 | 0.35 |
ENST00000380200.3
|
S100G
|
S100 calcium binding protein G |
| chr7_-_36634181 | 0.35 |
ENST00000538464.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr11_-_3663212 | 0.34 |
ENST00000397067.3
|
ART5
|
ADP-ribosyltransferase 5 |
| chr15_-_80263506 | 0.33 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr2_-_136678123 | 0.33 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr4_+_88754113 | 0.33 |
ENST00000560249.1
ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr4_-_76928641 | 0.32 |
ENST00000264888.5
|
CXCL9
|
chemokine (C-X-C motif) ligand 9 |
| chr5_+_148521381 | 0.31 |
ENST00000504238.1
|
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr7_+_141811539 | 0.31 |
ENST00000550469.2
ENST00000477922.3 |
RP11-1220K2.2
|
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr17_-_39306054 | 0.31 |
ENST00000343246.4
|
KRTAP4-5
|
keratin associated protein 4-5 |
| chr12_-_18243075 | 0.30 |
ENST00000536890.1
|
RERGL
|
RERG/RAS-like |
| chr12_+_6833437 | 0.29 |
ENST00000534947.1
ENST00000541866.1 ENST00000534877.1 ENST00000538753.1 |
COPS7A
|
COP9 signalosome subunit 7A |
| chr6_+_127898312 | 0.29 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr19_-_10420459 | 0.29 |
ENST00000403352.1
ENST00000403903.3 |
ZGLP1
|
zinc finger, GATA-like protein 1 |
| chr8_+_77318769 | 0.29 |
ENST00000518732.1
|
RP11-706J10.1
|
long intergenic non-protein coding RNA 1111 |
| chr6_-_82462425 | 0.29 |
ENST00000369754.3
ENST00000320172.6 ENST00000369756.3 |
FAM46A
|
family with sequence similarity 46, member A |
| chr12_+_6833237 | 0.29 |
ENST00000229251.3
ENST00000539735.1 ENST00000538410.1 |
COPS7A
|
COP9 signalosome subunit 7A |
| chr15_+_93749295 | 0.29 |
ENST00000599897.1
|
AC112693.2
|
AC112693.2 |
| chr22_+_21133469 | 0.29 |
ENST00000406799.1
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr19_-_53636125 | 0.29 |
ENST00000601493.1
ENST00000599261.1 ENST00000597503.1 ENST00000500065.4 ENST00000243643.4 ENST00000594011.1 ENST00000455735.2 ENST00000595193.1 ENST00000448501.1 ENST00000421033.1 ENST00000440291.1 ENST00000595813.1 ENST00000600574.1 ENST00000596051.1 ENST00000601110.1 |
ZNF415
|
zinc finger protein 415 |
| chr6_+_42584847 | 0.29 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr18_+_29769978 | 0.28 |
ENST00000269202.6
ENST00000581447.1 |
MEP1B
|
meprin A, beta |
| chr8_-_124749609 | 0.28 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr17_-_55911970 | 0.28 |
ENST00000581805.1
ENST00000580960.1 |
RP11-60A24.3
|
RP11-60A24.3 |
| chr18_-_19994830 | 0.28 |
ENST00000525417.1
|
CTAGE1
|
cutaneous T-cell lymphoma-associated antigen 1 |
| chr7_-_75401513 | 0.28 |
ENST00000005180.4
|
CCL26
|
chemokine (C-C motif) ligand 26 |
| chr3_-_150690471 | 0.28 |
ENST00000468836.1
ENST00000328863.4 |
CLRN1
|
clarin 1 |
| chr10_-_90751038 | 0.28 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr13_-_47471155 | 0.28 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr1_-_182369751 | 0.27 |
ENST00000367565.1
|
TEDDM1
|
transmembrane epididymal protein 1 |
| chr4_-_74964904 | 0.27 |
ENST00000508487.2
|
CXCL2
|
chemokine (C-X-C motif) ligand 2 |
| chr9_-_95640218 | 0.27 |
ENST00000395506.3
ENST00000375495.3 ENST00000332591.6 |
ZNF484
|
zinc finger protein 484 |
| chr20_+_37554955 | 0.27 |
ENST00000217429.4
|
FAM83D
|
family with sequence similarity 83, member D |
| chr6_+_12007963 | 0.27 |
ENST00000607445.1
|
RP11-456H18.2
|
RP11-456H18.2 |
| chr9_-_114521783 | 0.26 |
ENST00000394779.3
ENST00000394777.4 |
C9orf84
|
chromosome 9 open reading frame 84 |
| chrX_+_78003204 | 0.26 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr18_+_45778672 | 0.26 |
ENST00000600091.1
|
AC091150.1
|
HCG1818186; Uncharacterized protein |
| chr5_-_20575959 | 0.26 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr1_-_89664595 | 0.26 |
ENST00000355754.6
|
GBP4
|
guanylate binding protein 4 |
| chr7_-_69062391 | 0.26 |
ENST00000436600.2
|
RP5-942I16.1
|
RP5-942I16.1 |
| chr5_+_67586465 | 0.26 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr22_-_39190116 | 0.25 |
ENST00000406622.1
ENST00000216068.4 ENST00000406199.3 |
SUN2
DNAL4
|
Sad1 and UNC84 domain containing 2 dynein, axonemal, light chain 4 |
| chr17_+_51900239 | 0.25 |
ENST00000268919.4
|
KIF2B
|
kinesin family member 2B |
| chr9_+_125132803 | 0.25 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr10_+_85933494 | 0.25 |
ENST00000372126.3
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr11_-_117748138 | 0.25 |
ENST00000527717.1
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr3_-_151034734 | 0.24 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr11_-_5207612 | 0.23 |
ENST00000380367.1
|
OR52A1
|
olfactory receptor, family 52, subfamily A, member 1 |
| chr12_+_12878829 | 0.23 |
ENST00000326765.6
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr9_-_130829588 | 0.23 |
ENST00000373078.4
|
NAIF1
|
nuclear apoptosis inducing factor 1 |
| chr14_-_21567009 | 0.23 |
ENST00000556174.1
ENST00000554478.1 ENST00000553980.1 ENST00000421093.2 |
ZNF219
|
zinc finger protein 219 |
| chr8_+_31497271 | 0.23 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr10_+_91087651 | 0.23 |
ENST00000371818.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr7_+_141695633 | 0.23 |
ENST00000549489.2
|
MGAM
|
maltase-glucoamylase (alpha-glucosidase) |
| chr7_+_141695671 | 0.23 |
ENST00000497673.1
ENST00000475668.2 |
MGAM
|
maltase-glucoamylase (alpha-glucosidase) |
| chr15_-_85197501 | 0.23 |
ENST00000434634.2
|
WDR73
|
WD repeat domain 73 |
| chr4_-_95264008 | 0.23 |
ENST00000295256.5
|
HPGDS
|
hematopoietic prostaglandin D synthase |
| chr22_-_30642782 | 0.23 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr10_+_24497704 | 0.23 |
ENST00000376456.4
ENST00000458595.1 |
KIAA1217
|
KIAA1217 |
| chr2_+_89986318 | 0.22 |
ENST00000491977.1
|
IGKV2D-29
|
immunoglobulin kappa variable 2D-29 |
| chr6_-_119031228 | 0.22 |
ENST00000392500.3
ENST00000368488.5 ENST00000434604.1 |
CEP85L
|
centrosomal protein 85kDa-like |
| chr8_+_124084899 | 0.22 |
ENST00000287380.1
ENST00000309336.3 ENST00000519418.1 ENST00000327098.5 ENST00000522420.1 ENST00000521676.1 ENST00000378080.2 |
TBC1D31
|
TBC1 domain family, member 31 |
| chr16_+_57653989 | 0.22 |
ENST00000567835.1
ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr7_-_36764004 | 0.22 |
ENST00000431169.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr15_-_98417780 | 0.22 |
ENST00000503874.3
|
LINC00923
|
long intergenic non-protein coding RNA 923 |
| chr16_+_57653854 | 0.22 |
ENST00000568908.1
ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr14_-_61748550 | 0.21 |
ENST00000555868.1
|
TMEM30B
|
transmembrane protein 30B |
| chr10_-_129691195 | 0.21 |
ENST00000368671.3
|
CLRN3
|
clarin 3 |
| chr4_+_119810134 | 0.21 |
ENST00000434046.2
|
SYNPO2
|
synaptopodin 2 |
| chr1_+_120254510 | 0.21 |
ENST00000369409.4
|
PHGDH
|
phosphoglycerate dehydrogenase |
| chr15_-_54025300 | 0.21 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr4_+_71457970 | 0.21 |
ENST00000322937.6
|
AMBN
|
ameloblastin (enamel matrix protein) |
| chr3_-_57326704 | 0.20 |
ENST00000487349.1
ENST00000389601.3 |
ASB14
|
ankyrin repeat and SOCS box containing 14 |
| chr3_-_108672742 | 0.20 |
ENST00000261047.3
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr8_-_13372253 | 0.20 |
ENST00000316609.5
|
DLC1
|
deleted in liver cancer 1 |
| chr17_-_39203519 | 0.20 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr4_-_47983519 | 0.20 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr1_+_153004800 | 0.20 |
ENST00000392661.3
|
SPRR1B
|
small proline-rich protein 1B |
| chr15_-_99789736 | 0.20 |
ENST00000560235.1
ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr12_-_95941987 | 0.19 |
ENST00000537435.2
|
USP44
|
ubiquitin specific peptidase 44 |
| chr3_-_108672609 | 0.19 |
ENST00000393963.3
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr19_-_19626838 | 0.19 |
ENST00000360913.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr5_-_75008244 | 0.19 |
ENST00000510798.1
ENST00000446329.2 |
POC5
|
POC5 centriolar protein |
| chr9_-_21305312 | 0.19 |
ENST00000259555.4
|
IFNA5
|
interferon, alpha 5 |
| chr17_-_28257080 | 0.19 |
ENST00000579954.1
ENST00000540801.1 ENST00000269033.3 ENST00000590153.1 ENST00000582084.1 |
SSH2
|
slingshot protein phosphatase 2 |
| chr17_+_41132564 | 0.19 |
ENST00000361677.1
ENST00000589705.1 |
RUNDC1
|
RUN domain containing 1 |
| chr3_+_173116225 | 0.19 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr11_-_3663502 | 0.19 |
ENST00000359918.4
|
ART5
|
ADP-ribosyltransferase 5 |
| chr8_-_72274095 | 0.18 |
ENST00000303824.7
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr6_+_12717892 | 0.18 |
ENST00000379350.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr3_+_155838337 | 0.18 |
ENST00000490337.1
ENST00000389636.5 |
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr10_-_14050522 | 0.18 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr11_-_5345582 | 0.18 |
ENST00000328813.2
|
OR51B2
|
olfactory receptor, family 51, subfamily B, member 2 |
| chr11_-_3663480 | 0.18 |
ENST00000397068.3
|
ART5
|
ADP-ribosyltransferase 5 |
| chr10_+_90750493 | 0.18 |
ENST00000357339.2
ENST00000355279.2 |
FAS
|
Fas cell surface death receptor |
| chr6_-_26189304 | 0.18 |
ENST00000340756.2
|
HIST1H4D
|
histone cluster 1, H4d |
| chr11_+_45825616 | 0.18 |
ENST00000442528.2
ENST00000456334.1 ENST00000526817.1 |
SLC35C1
|
solute carrier family 35 (GDP-fucose transporter), member C1 |
| chr2_+_133874577 | 0.18 |
ENST00000596384.1
|
AC011755.1
|
HCG2006742; Protein LOC100996685 |
| chr5_-_160279207 | 0.18 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr11_+_35201826 | 0.18 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr9_+_77112244 | 0.17 |
ENST00000376896.3
|
RORB
|
RAR-related orphan receptor B |
| chr22_-_32651326 | 0.17 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr7_-_5553369 | 0.17 |
ENST00000453700.3
ENST00000382368.3 |
FBXL18
|
F-box and leucine-rich repeat protein 18 |
| chr19_+_41281282 | 0.17 |
ENST00000263369.3
|
MIA
|
melanoma inhibitory activity |
| chr19_+_1440838 | 0.17 |
ENST00000594262.1
|
AC027307.3
|
Uncharacterized protein |
| chr6_-_87804815 | 0.17 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chrX_+_144908928 | 0.17 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chrX_+_37865804 | 0.17 |
ENST00000297875.2
ENST00000357972.5 |
SYTL5
|
synaptotagmin-like 5 |
| chr1_-_106161540 | 0.17 |
ENST00000420901.1
ENST00000610126.1 ENST00000435253.2 |
RP11-251P6.1
|
RP11-251P6.1 |
| chr18_-_74728998 | 0.17 |
ENST00000359645.3
ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP
|
myelin basic protein |
| chr2_+_69201705 | 0.17 |
ENST00000377938.2
|
GKN1
|
gastrokine 1 |
| chr1_+_26485511 | 0.17 |
ENST00000374268.3
|
FAM110D
|
family with sequence similarity 110, member D |
| chr15_+_69857515 | 0.17 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr17_+_58018269 | 0.16 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr9_+_21967137 | 0.16 |
ENST00000441769.2
|
C9orf53
|
chromosome 9 open reading frame 53 |
| chr14_+_73563735 | 0.16 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25 |
| chr19_-_53770972 | 0.16 |
ENST00000311170.4
|
VN1R4
|
vomeronasal 1 receptor 4 |
| chr7_-_77045617 | 0.16 |
ENST00000257626.7
|
GSAP
|
gamma-secretase activating protein |
| chr1_+_241695424 | 0.16 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr4_-_68749699 | 0.16 |
ENST00000545541.1
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr10_-_62493223 | 0.16 |
ENST00000373827.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr12_-_49418407 | 0.16 |
ENST00000526209.1
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr12_+_82347498 | 0.16 |
ENST00000550506.1
|
RP11-362A1.1
|
RP11-362A1.1 |
| chr3_-_108476231 | 0.16 |
ENST00000295755.6
|
RETNLB
|
resistin like beta |
| chr12_-_30887948 | 0.16 |
ENST00000433722.2
|
CAPRIN2
|
caprin family member 2 |
| chr7_+_26591441 | 0.16 |
ENST00000420912.1
ENST00000457000.1 ENST00000430426.1 |
AC004947.2
|
AC004947.2 |
| chr1_+_23345943 | 0.16 |
ENST00000400181.4
ENST00000542151.1 |
KDM1A
|
lysine (K)-specific demethylase 1A |
| chr19_+_41281060 | 0.16 |
ENST00000594436.1
ENST00000597784.1 |
MIA
|
melanoma inhibitory activity |
| chr6_+_29274403 | 0.16 |
ENST00000377160.2
|
OR14J1
|
olfactory receptor, family 14, subfamily J, member 1 |
| chr1_+_23345930 | 0.16 |
ENST00000356634.3
|
KDM1A
|
lysine (K)-specific demethylase 1A |
| chr9_+_95736758 | 0.16 |
ENST00000337352.6
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr11_-_36619771 | 0.15 |
ENST00000311485.3
ENST00000527033.1 ENST00000532616.1 |
RAG2
|
recombination activating gene 2 |
| chr17_+_73717516 | 0.15 |
ENST00000200181.3
ENST00000339591.3 |
ITGB4
|
integrin, beta 4 |
| chr15_-_50558223 | 0.15 |
ENST00000267845.3
|
HDC
|
histidine decarboxylase |
| chr10_+_17794251 | 0.15 |
ENST00000377495.1
ENST00000338221.5 |
TMEM236
|
transmembrane protein 236 |
| chr12_+_70219052 | 0.15 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chr1_-_237167718 | 0.15 |
ENST00000464121.2
|
MT1HL1
|
metallothionein 1H-like 1 |
| chr1_-_68962805 | 0.15 |
ENST00000370966.5
|
DEPDC1
|
DEP domain containing 1 |
| chr6_-_9939552 | 0.15 |
ENST00000460363.2
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr6_+_158733692 | 0.15 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr10_-_29084886 | 0.15 |
ENST00000608061.1
ENST00000443246.2 ENST00000446012.1 |
LINC00837
|
long intergenic non-protein coding RNA 837 |
| chr13_-_103719196 | 0.15 |
ENST00000245312.3
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr12_-_6579808 | 0.15 |
ENST00000535180.1
ENST00000400911.3 |
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr4_-_84035905 | 0.15 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr6_-_22297730 | 0.15 |
ENST00000306482.1
|
PRL
|
prolactin |
| chr16_-_30381580 | 0.15 |
ENST00000409939.3
|
TBC1D10B
|
TBC1 domain family, member 10B |
| chr12_-_101604185 | 0.15 |
ENST00000536262.2
|
SLC5A8
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 8 |
| chr7_+_73106926 | 0.15 |
ENST00000453316.1
|
WBSCR22
|
Williams Beuren syndrome chromosome region 22 |
| chr2_-_190044480 | 0.15 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr6_+_12007897 | 0.14 |
ENST00000437559.1
|
RP11-456H18.2
|
RP11-456H18.2 |
| chr10_+_96522361 | 0.14 |
ENST00000371321.3
|
CYP2C19
|
cytochrome P450, family 2, subfamily C, polypeptide 19 |
| chr5_-_59783882 | 0.14 |
ENST00000505507.2
ENST00000502484.2 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD11_HOXA11
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.3 | 0.8 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.4 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 1.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.7 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.3 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.4 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.4 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.1 | 0.5 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.1 | 0.1 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 0.2 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.1 | 0.2 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.1 | 0.2 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.1 | 0.7 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.2 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.7 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 0.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.4 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.3 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.3 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.1 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.2 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.4 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.1 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.6 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.2 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.0 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0032900 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) negative regulation of neurotrophin production(GO:0032900) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.2 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.0 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.2 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.0 | GO:1902908 | regulation of melanosome transport(GO:1902908) |
| 0.0 | 0.2 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.4 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.3 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.2 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.2 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.0 | 0.6 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 4.7 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.2 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.5 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.1 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.4 | GO:0070168 | negative regulation of biomineral tissue development(GO:0070168) |
| 0.0 | 0.1 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.0 | GO:1902771 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.0 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.3 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.3 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.1 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.0 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 1.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.5 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.0 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.0 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) response to fluoride(GO:1902617) |
| 0.0 | 1.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.0 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.2 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 0.3 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.2 | 1.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.7 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 0.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 4.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.6 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 1.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 0.6 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 1.0 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.7 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 0.5 | GO:0016160 | amylase activity(GO:0016160) |
| 0.1 | 0.3 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.1 | 1.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.4 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.4 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.1 | 0.5 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 0.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.6 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.1 | 0.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 0.3 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 0.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.3 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 1.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.2 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.1 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.1 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.2 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.5 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.4 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.3 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0060230 | lipase binding(GO:0035473) lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.0 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.0 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 1.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.0 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.4 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.0 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 2.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |