Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
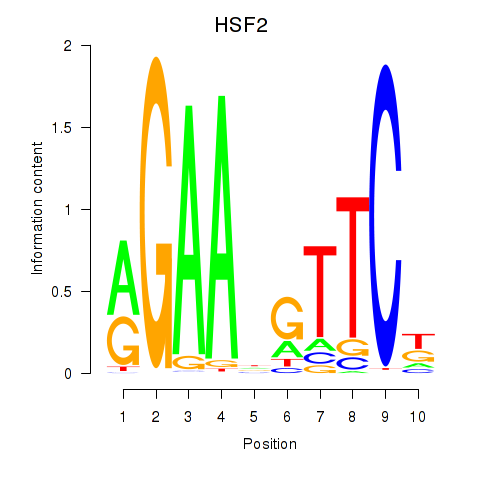
Results for HSF2
Z-value: 0.65
Transcription factors associated with HSF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF2
|
ENSG00000025156.8 | heat shock transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSF2 | hg19_v2_chr6_+_122720681_122720713 | -0.18 | 3.8e-01 | Click! |
Activity profile of HSF2 motif
Sorted Z-values of HSF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_30960915 | 1.51 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr8_-_81083341 | 1.30 |
ENST00000519303.2
|
TPD52
|
tumor protein D52 |
| chr6_+_149539767 | 1.11 |
ENST00000606202.1
ENST00000536230.1 ENST00000445901.1 |
TAB2
RP1-111D6.3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 RP1-111D6.3 |
| chr8_-_81083890 | 0.95 |
ENST00000518937.1
|
TPD52
|
tumor protein D52 |
| chr4_-_186877806 | 0.88 |
ENST00000355634.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_+_118691706 | 0.79 |
ENST00000415806.2
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr4_-_186877502 | 0.71 |
ENST00000431902.1
ENST00000284776.7 ENST00000415274.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chrX_+_135279179 | 0.70 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr8_-_81083731 | 0.70 |
ENST00000379096.5
|
TPD52
|
tumor protein D52 |
| chr9_+_33025209 | 0.65 |
ENST00000330899.4
ENST00000544625.1 |
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1 |
| chrX_+_135230712 | 0.63 |
ENST00000535737.1
|
FHL1
|
four and a half LIM domains 1 |
| chr15_-_70388599 | 0.59 |
ENST00000560996.1
ENST00000558201.1 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr19_-_14628645 | 0.58 |
ENST00000598235.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chrX_+_135278908 | 0.57 |
ENST00000539015.1
ENST00000370683.1 |
FHL1
|
four and a half LIM domains 1 |
| chr7_+_75931861 | 0.56 |
ENST00000248553.6
|
HSPB1
|
heat shock 27kDa protein 1 |
| chr11_+_12695944 | 0.53 |
ENST00000361905.4
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr15_-_70388943 | 0.53 |
ENST00000559048.1
ENST00000560939.1 ENST00000440567.3 ENST00000557907.1 ENST00000558379.1 ENST00000451782.2 ENST00000559929.1 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr5_+_56469843 | 0.51 |
ENST00000514387.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr2_-_188419078 | 0.51 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr12_-_123011476 | 0.50 |
ENST00000528279.1
ENST00000344591.4 ENST00000526560.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr18_-_52989217 | 0.49 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr17_+_56315936 | 0.46 |
ENST00000543544.1
|
LPO
|
lactoperoxidase |
| chr5_+_56469939 | 0.46 |
ENST00000506184.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr5_+_56469775 | 0.45 |
ENST00000424459.3
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr2_-_188419200 | 0.44 |
ENST00000233156.3
ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr12_-_10007448 | 0.43 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr11_+_46316677 | 0.42 |
ENST00000534787.1
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1 |
| chr12_-_14721283 | 0.42 |
ENST00000240617.5
|
PLBD1
|
phospholipase B domain containing 1 |
| chr11_+_117063295 | 0.42 |
ENST00000525478.1
ENST00000532062.1 |
SIDT2
|
SID1 transmembrane family, member 2 |
| chr11_-_71791518 | 0.41 |
ENST00000537217.1
ENST00000366394.3 ENST00000358965.6 ENST00000546131.1 ENST00000543937.1 ENST00000368959.5 ENST00000541641.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr7_+_130126165 | 0.41 |
ENST00000427521.1
ENST00000416162.2 ENST00000378576.4 |
MEST
|
mesoderm specific transcript |
| chr7_+_130126012 | 0.40 |
ENST00000341441.5
|
MEST
|
mesoderm specific transcript |
| chr11_-_71791726 | 0.40 |
ENST00000393695.3
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr3_-_64673668 | 0.39 |
ENST00000498707.1
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chr17_-_79817091 | 0.37 |
ENST00000570907.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chrX_-_30993201 | 0.36 |
ENST00000288422.2
ENST00000378932.2 |
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr18_-_812231 | 0.36 |
ENST00000314574.4
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr12_+_2904102 | 0.33 |
ENST00000001008.4
|
FKBP4
|
FK506 binding protein 4, 59kDa |
| chr3_-_112360116 | 0.33 |
ENST00000206423.3
ENST00000439685.2 |
CCDC80
|
coiled-coil domain containing 80 |
| chr6_+_44215603 | 0.33 |
ENST00000371554.1
|
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr12_+_69979210 | 0.32 |
ENST00000544368.2
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr6_+_29364416 | 0.32 |
ENST00000383555.2
|
OR12D2
|
olfactory receptor, family 12, subfamily D, member 2 (gene/pseudogene) |
| chr5_-_112770674 | 0.31 |
ENST00000390666.3
|
TSSK1B
|
testis-specific serine kinase 1B |
| chr18_-_52989525 | 0.31 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr6_+_31783291 | 0.30 |
ENST00000375651.5
ENST00000608703.1 ENST00000458062.2 |
HSPA1A
|
heat shock 70kDa protein 1A |
| chr7_+_23637763 | 0.29 |
ENST00000410069.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr2_-_85581623 | 0.28 |
ENST00000449375.1
ENST00000409984.2 ENST00000457495.2 ENST00000263854.6 |
RETSAT
|
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr2_-_178128250 | 0.28 |
ENST00000448782.1
ENST00000446151.2 |
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr4_-_151936416 | 0.28 |
ENST00000510413.1
ENST00000507224.1 |
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr12_-_123011536 | 0.27 |
ENST00000331738.7
ENST00000354654.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr4_-_6383594 | 0.27 |
ENST00000335585.5
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr2_+_28615669 | 0.26 |
ENST00000379619.1
ENST00000264716.4 |
FOSL2
|
FOS-like antigen 2 |
| chr5_+_79331164 | 0.26 |
ENST00000350881.2
|
THBS4
|
thrombospondin 4 |
| chr19_-_14629224 | 0.26 |
ENST00000254322.2
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr2_+_74648848 | 0.25 |
ENST00000409791.1
ENST00000426787.1 ENST00000348227.4 |
WDR54
|
WD repeat domain 54 |
| chr4_-_22517620 | 0.25 |
ENST00000502482.1
ENST00000334304.5 |
GPR125
|
G protein-coupled receptor 125 |
| chr14_+_103800513 | 0.25 |
ENST00000560338.1
ENST00000560763.1 ENST00000558316.1 ENST00000558265.1 |
EIF5
|
eukaryotic translation initiation factor 5 |
| chr12_+_69979113 | 0.24 |
ENST00000299300.6
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr12_+_11802753 | 0.24 |
ENST00000396373.4
|
ETV6
|
ets variant 6 |
| chr3_-_183735651 | 0.24 |
ENST00000427120.2
ENST00000392579.2 ENST00000382494.2 ENST00000446941.2 |
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr15_+_65134088 | 0.23 |
ENST00000323544.4
ENST00000437723.1 |
PLEKHO2
AC069368.3
|
pleckstrin homology domain containing, family O member 2 Uncharacterized protein |
| chr12_+_69979446 | 0.22 |
ENST00000543146.2
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr8_+_134125727 | 0.22 |
ENST00000521107.1
|
TG
|
thyroglobulin |
| chr11_+_86013253 | 0.22 |
ENST00000533986.1
ENST00000278483.3 |
C11orf73
|
chromosome 11 open reading frame 73 |
| chr11_-_414948 | 0.22 |
ENST00000530494.1
ENST00000528209.1 ENST00000431843.2 ENST00000528058.1 |
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr7_-_4901625 | 0.22 |
ENST00000404991.1
|
PAPOLB
|
poly(A) polymerase beta (testis specific) |
| chr2_-_97405775 | 0.22 |
ENST00000264963.4
ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L
|
lectin, mannose-binding 2-like |
| chrX_-_107225600 | 0.21 |
ENST00000302917.1
|
TEX13B
|
testis expressed 13B |
| chr5_-_159739532 | 0.21 |
ENST00000520748.1
ENST00000393977.3 ENST00000257536.7 |
CCNJL
|
cyclin J-like |
| chr6_+_32709119 | 0.21 |
ENST00000374940.3
|
HLA-DQA2
|
major histocompatibility complex, class II, DQ alpha 2 |
| chr2_-_85581701 | 0.21 |
ENST00000295802.4
|
RETSAT
|
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr9_+_91926103 | 0.21 |
ENST00000314355.6
|
CKS2
|
CDC28 protein kinase regulatory subunit 2 |
| chr6_-_155635583 | 0.21 |
ENST00000367166.4
|
TFB1M
|
transcription factor B1, mitochondrial |
| chr6_-_53013620 | 0.20 |
ENST00000259803.7
|
GCM1
|
glial cells missing homolog 1 (Drosophila) |
| chr2_+_219824357 | 0.20 |
ENST00000302625.4
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39) |
| chr13_+_24144796 | 0.19 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr19_-_49522727 | 0.19 |
ENST00000600007.1
|
CTB-60B18.10
|
CTB-60B18.10 |
| chr17_-_74137374 | 0.18 |
ENST00000322957.6
|
FOXJ1
|
forkhead box J1 |
| chr13_+_24144509 | 0.18 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr7_-_26240357 | 0.18 |
ENST00000354667.4
ENST00000356674.7 |
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr13_+_46276441 | 0.17 |
ENST00000310521.1
ENST00000533564.1 |
SPERT
|
spermatid associated |
| chr20_+_57427765 | 0.17 |
ENST00000371100.4
|
GNAS
|
GNAS complex locus |
| chr7_-_33080506 | 0.17 |
ENST00000381626.2
ENST00000409467.1 ENST00000449201.1 |
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr17_-_76573465 | 0.17 |
ENST00000585328.1
ENST00000389840.5 |
DNAH17
|
dynein, axonemal, heavy chain 17 |
| chr14_+_89060749 | 0.16 |
ENST00000555900.1
ENST00000406216.3 ENST00000557737.1 |
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr15_+_36338242 | 0.16 |
ENST00000560056.1
|
RP11-684B21.1
|
RP11-684B21.1 |
| chr12_-_45269430 | 0.16 |
ENST00000395487.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr10_+_120116527 | 0.16 |
ENST00000445161.1
|
LINC00867
|
long intergenic non-protein coding RNA 867 |
| chr17_-_56494908 | 0.16 |
ENST00000577716.1
|
RNF43
|
ring finger protein 43 |
| chr13_-_31736132 | 0.16 |
ENST00000429785.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr6_+_106534192 | 0.16 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr7_-_14880892 | 0.15 |
ENST00000406247.3
ENST00000399322.3 ENST00000258767.5 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chrX_-_51239425 | 0.15 |
ENST00000375992.3
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr11_+_65770227 | 0.15 |
ENST00000527348.1
|
BANF1
|
barrier to autointegration factor 1 |
| chr9_+_34989638 | 0.15 |
ENST00000453597.3
ENST00000335998.3 ENST00000312316.5 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr17_-_56494882 | 0.15 |
ENST00000584437.1
|
RNF43
|
ring finger protein 43 |
| chr14_+_89060739 | 0.15 |
ENST00000318308.6
|
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr7_+_132333553 | 0.14 |
ENST00000332558.4
|
AC009365.3
|
AC009365.3 |
| chr11_+_12696102 | 0.14 |
ENST00000527636.1
ENST00000527376.1 |
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr15_-_43785274 | 0.14 |
ENST00000413546.1
|
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr19_-_49560987 | 0.14 |
ENST00000596965.1
|
CGB7
|
chorionic gonadotropin, beta polypeptide 7 |
| chr17_-_56494713 | 0.13 |
ENST00000407977.2
|
RNF43
|
ring finger protein 43 |
| chr21_+_35445811 | 0.13 |
ENST00000399312.2
|
MRPS6
|
mitochondrial ribosomal protein S6 |
| chr12_-_45270077 | 0.13 |
ENST00000551601.1
ENST00000549027.1 ENST00000452445.2 |
NELL2
|
NEL-like 2 (chicken) |
| chr3_+_39509070 | 0.13 |
ENST00000354668.4
ENST00000428261.1 ENST00000420739.1 ENST00000415443.1 ENST00000447324.1 ENST00000383754.3 |
MOBP
|
myelin-associated oligodendrocyte basic protein |
| chr11_+_65769946 | 0.13 |
ENST00000533166.1
|
BANF1
|
barrier to autointegration factor 1 |
| chr4_+_123161120 | 0.13 |
ENST00000446180.1
|
KIAA1109
|
KIAA1109 |
| chr13_-_31736478 | 0.13 |
ENST00000445273.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr20_+_4666882 | 0.13 |
ENST00000379440.4
ENST00000430350.2 |
PRNP
|
prion protein |
| chr7_+_142378566 | 0.12 |
ENST00000390398.3
|
TRBV25-1
|
T cell receptor beta variable 25-1 |
| chr18_+_3451584 | 0.12 |
ENST00000551541.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr2_-_10978103 | 0.12 |
ENST00000404824.2
|
PDIA6
|
protein disulfide isomerase family A, member 6 |
| chr20_+_31755934 | 0.12 |
ENST00000354932.5
|
BPIFA2
|
BPI fold containing family A, member 2 |
| chr20_+_61867235 | 0.12 |
ENST00000342412.6
ENST00000217169.3 |
BIRC7
|
baculoviral IAP repeat containing 7 |
| chr11_+_65769550 | 0.12 |
ENST00000312175.2
ENST00000445560.2 ENST00000530204.1 |
BANF1
|
barrier to autointegration factor 1 |
| chr2_-_197226875 | 0.12 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr22_+_40297079 | 0.11 |
ENST00000344138.4
ENST00000543252.1 |
GRAP2
|
GRB2-related adaptor protein 2 |
| chr11_+_66276550 | 0.11 |
ENST00000419755.3
|
CTD-3074O7.11
|
Bardet-Biedl syndrome 1 protein |
| chr18_+_3451646 | 0.11 |
ENST00000345133.5
ENST00000330513.5 ENST00000549546.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr8_-_141931287 | 0.11 |
ENST00000517887.1
|
PTK2
|
protein tyrosine kinase 2 |
| chr6_-_53213780 | 0.11 |
ENST00000304434.6
ENST00000370918.4 |
ELOVL5
|
ELOVL fatty acid elongase 5 |
| chr9_-_4666337 | 0.11 |
ENST00000381890.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr4_-_159644507 | 0.11 |
ENST00000307720.3
|
PPID
|
peptidylprolyl isomerase D |
| chr2_-_152589670 | 0.10 |
ENST00000604864.1
ENST00000603639.1 |
NEB
|
nebulin |
| chr17_+_7608511 | 0.10 |
ENST00000226091.2
|
EFNB3
|
ephrin-B3 |
| chr12_+_54891495 | 0.10 |
ENST00000293373.6
|
NCKAP1L
|
NCK-associated protein 1-like |
| chr19_+_54606145 | 0.10 |
ENST00000485876.1
ENST00000391762.1 ENST00000471292.1 ENST00000391763.3 ENST00000391764.3 ENST00000303553.5 |
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa |
| chr22_+_40297105 | 0.10 |
ENST00000540310.1
|
GRAP2
|
GRB2-related adaptor protein 2 |
| chr3_-_183735731 | 0.10 |
ENST00000334444.6
|
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr3_-_36781352 | 0.10 |
ENST00000416516.2
|
DCLK3
|
doublecortin-like kinase 3 |
| chr17_+_73663402 | 0.09 |
ENST00000355423.3
|
SAP30BP
|
SAP30 binding protein |
| chr9_-_4666421 | 0.09 |
ENST00000381895.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr1_+_192544857 | 0.09 |
ENST00000367459.3
ENST00000469578.2 |
RGS1
|
regulator of G-protein signaling 1 |
| chr14_-_57277178 | 0.09 |
ENST00000339475.5
ENST00000554559.1 ENST00000555804.1 |
OTX2
|
orthodenticle homeobox 2 |
| chr4_+_154387480 | 0.08 |
ENST00000409663.3
ENST00000440693.1 ENST00000409959.3 |
KIAA0922
|
KIAA0922 |
| chr20_+_33292068 | 0.08 |
ENST00000374810.3
ENST00000374809.2 ENST00000451665.1 |
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr6_+_32121218 | 0.08 |
ENST00000414204.1
ENST00000361568.2 ENST00000395523.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr6_-_32339649 | 0.08 |
ENST00000527965.1
ENST00000532023.1 ENST00000447241.2 ENST00000375007.4 ENST00000534588.1 |
C6orf10
|
chromosome 6 open reading frame 10 |
| chr1_+_168195229 | 0.07 |
ENST00000271375.4
ENST00000367825.3 |
SFT2D2
|
SFT2 domain containing 2 |
| chr6_-_32339671 | 0.07 |
ENST00000442822.2
ENST00000375015.4 ENST00000533191.1 |
C6orf10
|
chromosome 6 open reading frame 10 |
| chr15_+_78556428 | 0.07 |
ENST00000394855.3
ENST00000489435.2 |
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr7_+_120629653 | 0.07 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr11_-_122932730 | 0.07 |
ENST00000532182.1
ENST00000524590.1 ENST00000528292.1 ENST00000533540.1 ENST00000525463.1 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr1_-_168698433 | 0.07 |
ENST00000367817.3
|
DPT
|
dermatopontin |
| chr10_-_1246317 | 0.07 |
ENST00000381305.1
|
ADARB2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr10_+_104535994 | 0.06 |
ENST00000369889.4
|
WBP1L
|
WW domain binding protein 1-like |
| chr8_-_120605194 | 0.06 |
ENST00000522167.1
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr11_-_82997420 | 0.06 |
ENST00000455220.2
ENST00000529689.1 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr11_+_75273246 | 0.06 |
ENST00000526397.1
ENST00000529643.1 ENST00000525492.1 ENST00000530284.1 ENST00000532356.1 ENST00000524558.1 |
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
| chr11_+_75273101 | 0.06 |
ENST00000533603.1
ENST00000358171.3 ENST00000526242.1 |
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
| chrX_-_101718085 | 0.06 |
ENST00000372750.1
ENST00000372752.1 |
NXF2B
|
nuclear RNA export factor 2B |
| chr5_-_35089722 | 0.05 |
ENST00000511486.1
ENST00000310101.5 ENST00000231423.3 ENST00000513753.1 ENST00000348262.3 ENST00000397391.3 ENST00000542609.1 |
PRLR
|
prolactin receptor |
| chr13_-_20805109 | 0.05 |
ENST00000241124.6
|
GJB6
|
gap junction protein, beta 6, 30kDa |
| chr11_-_65769594 | 0.05 |
ENST00000532707.1
ENST00000533544.1 ENST00000526451.1 ENST00000312234.2 ENST00000530462.1 ENST00000525767.1 ENST00000529964.1 ENST00000527249.1 |
EIF1AD
|
eukaryotic translation initiation factor 1A domain containing |
| chr15_-_81616446 | 0.05 |
ENST00000302824.6
|
STARD5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr21_-_30445886 | 0.05 |
ENST00000431234.1
ENST00000540844.1 ENST00000286788.4 |
CCT8
|
chaperonin containing TCP1, subunit 8 (theta) |
| chr3_+_158787041 | 0.05 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr21_+_45148735 | 0.05 |
ENST00000327574.4
|
PDXK
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr14_+_93357749 | 0.05 |
ENST00000557689.1
|
RP11-862G15.1
|
RP11-862G15.1 |
| chr17_-_71410794 | 0.05 |
ENST00000424778.1
|
SDK2
|
sidekick cell adhesion molecule 2 |
| chr19_+_1438383 | 0.05 |
ENST00000586686.2
ENST00000591032.1 ENST00000586656.1 ENST00000589656.2 ENST00000593052.1 ENST00000586096.2 ENST00000591804.2 |
RPS15
|
ribosomal protein S15 |
| chr6_+_41303372 | 0.04 |
ENST00000373083.4
ENST00000373089.5 ENST00000373086.3 |
NCR2
|
natural cytotoxicity triggering receptor 2 |
| chr14_-_25479811 | 0.04 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr19_-_55691614 | 0.04 |
ENST00000592470.1
ENST00000354308.3 |
SYT5
|
synaptotagmin V |
| chr13_-_31736027 | 0.04 |
ENST00000380406.5
ENST00000320027.5 ENST00000380405.4 |
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr6_+_32121789 | 0.04 |
ENST00000437001.2
ENST00000375137.2 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr18_+_3449821 | 0.04 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr19_+_1438351 | 0.04 |
ENST00000233609.4
|
RPS15
|
ribosomal protein S15 |
| chrX_+_101478829 | 0.04 |
ENST00000372763.1
ENST00000372758.1 |
NXF2
|
nuclear RNA export factor 2 |
| chr1_-_228613026 | 0.04 |
ENST00000366696.1
|
HIST3H3
|
histone cluster 3, H3 |
| chr6_+_32121908 | 0.04 |
ENST00000375143.2
ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr2_+_198365122 | 0.03 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr3_+_96533413 | 0.03 |
ENST00000470610.2
ENST00000389672.5 |
EPHA6
|
EPH receptor A6 |
| chr6_+_31795506 | 0.03 |
ENST00000375650.3
|
HSPA1B
|
heat shock 70kDa protein 1B |
| chr15_+_78556809 | 0.03 |
ENST00000343789.3
ENST00000394852.3 |
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr19_+_782755 | 0.03 |
ENST00000606242.1
ENST00000586061.1 |
AC006273.5
|
AC006273.5 |
| chr6_-_123957942 | 0.03 |
ENST00000398178.3
|
TRDN
|
triadin |
| chr3_-_49131013 | 0.03 |
ENST00000424300.1
|
QRICH1
|
glutamine-rich 1 |
| chr10_+_135340859 | 0.03 |
ENST00000252945.3
ENST00000421586.1 ENST00000418356.1 |
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr15_+_96869165 | 0.03 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr12_+_58138800 | 0.02 |
ENST00000547992.1
ENST00000552816.1 ENST00000547472.1 |
TSPAN31
|
tetraspanin 31 |
| chr3_+_39509163 | 0.02 |
ENST00000436143.2
ENST00000441980.2 ENST00000311042.6 |
MOBP
|
myelin-associated oligodendrocyte basic protein |
| chr8_+_87878640 | 0.02 |
ENST00000518476.1
|
CNBD1
|
cyclic nucleotide binding domain containing 1 |
| chr7_-_150652924 | 0.02 |
ENST00000330883.4
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr6_-_123958141 | 0.02 |
ENST00000334268.4
|
TRDN
|
triadin |
| chr11_+_27062502 | 0.02 |
ENST00000263182.3
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr1_-_216596738 | 0.01 |
ENST00000307340.3
ENST00000366943.2 ENST00000366942.3 |
USH2A
|
Usher syndrome 2A (autosomal recessive, mild) |
| chr3_-_185655795 | 0.01 |
ENST00000342294.4
ENST00000382191.4 ENST00000453386.2 |
TRA2B
|
transformer 2 beta homolog (Drosophila) |
| chr16_+_55600580 | 0.01 |
ENST00000457326.2
|
CAPNS2
|
calpain, small subunit 2 |
| chrX_+_106163626 | 0.01 |
ENST00000336803.1
|
CLDN2
|
claudin 2 |
| chr19_+_14142535 | 0.01 |
ENST00000263379.2
|
IL27RA
|
interleukin 27 receptor, alpha |
| chr3_-_48632593 | 0.01 |
ENST00000454817.1
ENST00000328333.8 |
COL7A1
|
collagen, type VII, alpha 1 |
| chr1_+_151171012 | 0.01 |
ENST00000349792.5
ENST00000409426.1 ENST00000441902.2 ENST00000368890.4 ENST00000424999.1 ENST00000368888.4 |
PIP5K1A
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha |
| chr4_-_107957454 | 0.01 |
ENST00000285311.3
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr11_+_27062860 | 0.01 |
ENST00000528583.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr19_-_55691472 | 0.00 |
ENST00000537500.1
|
SYT5
|
synaptotagmin V |
| chrX_-_102941596 | 0.00 |
ENST00000441076.2
ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr11_+_71791693 | 0.00 |
ENST00000289488.2
ENST00000447974.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr14_-_94970559 | 0.00 |
ENST00000556881.1
|
SERPINA12
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
| chr14_-_77889860 | 0.00 |
ENST00000555603.1
|
NOXRED1
|
NADP-dependent oxidoreductase domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HSF2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.3 | 0.8 | GO:0032887 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.3 | 0.8 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.1 | 0.6 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.1 | 0.9 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 1.0 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.1 | 0.4 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.3 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.1 | 0.3 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 0.2 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.2 | GO:0002661 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.1 | 0.8 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.3 | GO:0046223 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.1 | 0.3 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 0.2 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.4 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 1.3 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.4 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.2 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:1990535 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) neuron projection maintenance(GO:1990535) |
| 0.0 | 0.2 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) regulation of NK T cell differentiation(GO:0051136) |
| 0.0 | 0.4 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 1.9 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 1.5 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.7 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.4 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.3 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.3 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 1.1 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 3.0 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.2 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.0 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 0.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.4 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.3 | 0.8 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.1 | 0.7 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 0.4 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.7 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.7 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.1 | 0.6 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.1 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.3 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.3 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.1 | 1.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 0.4 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.2 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 0.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.4 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.3 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 1.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.7 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.3 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.0 | GO:0031403 | pyridoxal kinase activity(GO:0008478) lithium ion binding(GO:0031403) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 2.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.8 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 3.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.9 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |