Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for HSF4
Z-value: 0.47
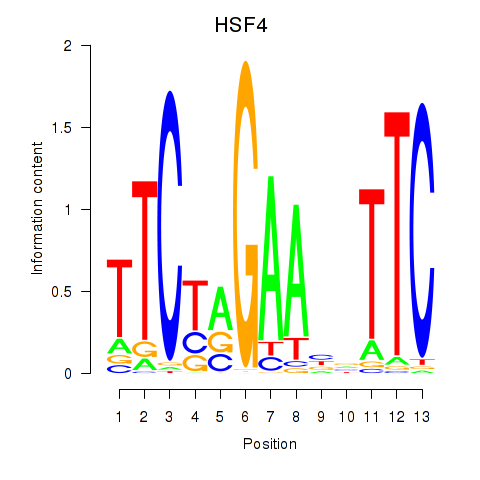
Transcription factors associated with HSF4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF4
|
ENSG00000102878.11 | heat shock transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSF4 | hg19_v2_chr16_+_67198683_67198715 | 0.04 | 8.7e-01 | Click! |
Activity profile of HSF4 motif
Sorted Z-values of HSF4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_56118981 | 1.44 |
ENST00000419984.2
ENST00000413218.1 ENST00000424596.1 |
PSPH
|
phosphoserine phosphatase |
| chr7_-_56119156 | 1.41 |
ENST00000421312.1
ENST00000416592.1 |
PSPH
|
phosphoserine phosphatase |
| chr21_-_31744557 | 1.18 |
ENST00000399889.2
|
KRTAP13-2
|
keratin associated protein 13-2 |
| chr2_+_228678550 | 0.68 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr11_-_111782484 | 0.51 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr4_-_84406218 | 0.38 |
ENST00000515303.1
|
FAM175A
|
family with sequence similarity 175, member A |
| chr13_-_31736132 | 0.36 |
ENST00000429785.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr1_-_60539405 | 0.36 |
ENST00000450089.2
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr1_-_60539422 | 0.36 |
ENST00000371201.3
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr2_-_100759037 | 0.34 |
ENST00000317233.4
ENST00000423966.1 ENST00000416492.1 |
AFF3
|
AF4/FMR2 family, member 3 |
| chrX_+_108779870 | 0.28 |
ENST00000372107.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr2_+_201170703 | 0.28 |
ENST00000358677.5
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr19_+_45971246 | 0.27 |
ENST00000585836.1
ENST00000417353.2 ENST00000353609.3 ENST00000591858.1 ENST00000443841.2 ENST00000590335.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr11_-_111782696 | 0.27 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr3_+_101443476 | 0.27 |
ENST00000327230.4
ENST00000494050.1 |
CEP97
|
centrosomal protein 97kDa |
| chr11_-_111794446 | 0.27 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr7_-_56119238 | 0.24 |
ENST00000275605.3
ENST00000395471.3 |
PSPH
|
phosphoserine phosphatase |
| chr10_-_74856608 | 0.24 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chr14_-_52535712 | 0.24 |
ENST00000216286.5
ENST00000541773.1 |
NID2
|
nidogen 2 (osteonidogen) |
| chr20_+_18794370 | 0.23 |
ENST00000377428.2
|
SCP2D1
|
SCP2 sterol-binding domain containing 1 |
| chr18_-_76738843 | 0.23 |
ENST00000575722.1
ENST00000583511.1 |
RP11-849I19.1
|
RP11-849I19.1 |
| chr8_+_143916217 | 0.23 |
ENST00000220940.1
|
GML
|
glycosylphosphatidylinositol anchored molecule like |
| chrX_-_154563889 | 0.22 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr20_-_60573188 | 0.22 |
ENST00000474089.1
|
TAF4
|
TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa |
| chr15_-_58357866 | 0.22 |
ENST00000537372.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr13_-_31736478 | 0.22 |
ENST00000445273.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr18_-_71815051 | 0.21 |
ENST00000582526.1
ENST00000419743.2 |
FBXO15
|
F-box protein 15 |
| chr18_-_71814999 | 0.21 |
ENST00000269500.5
|
FBXO15
|
F-box protein 15 |
| chr9_-_137809718 | 0.20 |
ENST00000371806.3
|
FCN1
|
ficolin (collagen/fibrinogen domain containing) 1 |
| chr7_-_3083573 | 0.20 |
ENST00000396946.4
|
CARD11
|
caspase recruitment domain family, member 11 |
| chr11_-_111781610 | 0.20 |
ENST00000525823.1
|
CRYAB
|
crystallin, alpha B |
| chr11_-_111781454 | 0.20 |
ENST00000533280.1
|
CRYAB
|
crystallin, alpha B |
| chr11_-_111781554 | 0.19 |
ENST00000526167.1
ENST00000528961.1 |
CRYAB
|
crystallin, alpha B |
| chr19_+_41509851 | 0.19 |
ENST00000593831.1
ENST00000330446.5 |
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr12_-_48500085 | 0.19 |
ENST00000549518.1
|
SENP1
|
SUMO1/sentrin specific peptidase 1 |
| chr19_-_49565254 | 0.19 |
ENST00000593537.1
|
NTF4
|
neurotrophin 4 |
| chr5_+_159895275 | 0.18 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr3_-_36781352 | 0.18 |
ENST00000416516.2
|
DCLK3
|
doublecortin-like kinase 3 |
| chr2_+_223162866 | 0.17 |
ENST00000295226.1
|
CCDC140
|
coiled-coil domain containing 140 |
| chrX_-_39923656 | 0.17 |
ENST00000413905.1
|
BCOR
|
BCL6 corepressor |
| chr11_+_107799118 | 0.17 |
ENST00000320578.2
|
RAB39A
|
RAB39A, member RAS oncogene family |
| chr21_+_44394742 | 0.16 |
ENST00000432907.2
|
PKNOX1
|
PBX/knotted 1 homeobox 1 |
| chr8_+_37594185 | 0.16 |
ENST00000518586.1
ENST00000335171.6 ENST00000521644.1 |
ERLIN2
|
ER lipid raft associated 2 |
| chr22_+_48972118 | 0.16 |
ENST00000358295.5
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5 |
| chr19_-_17958832 | 0.16 |
ENST00000458235.1
|
JAK3
|
Janus kinase 3 |
| chr2_-_69180012 | 0.15 |
ENST00000481498.1
|
GKN2
|
gastrokine 2 |
| chr11_+_67007518 | 0.14 |
ENST00000530342.1
ENST00000308783.5 |
KDM2A
|
lysine (K)-specific demethylase 2A |
| chr13_-_31736027 | 0.14 |
ENST00000380406.5
ENST00000320027.5 ENST00000380405.4 |
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chrX_-_152939252 | 0.14 |
ENST00000340888.3
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr17_+_74722912 | 0.13 |
ENST00000589977.1
ENST00000591571.1 ENST00000592849.1 ENST00000586738.1 ENST00000588783.1 ENST00000588563.1 ENST00000586752.1 ENST00000588302.1 ENST00000590964.1 ENST00000341249.6 ENST00000588822.1 |
METTL23
|
methyltransferase like 23 |
| chr15_-_89755034 | 0.13 |
ENST00000563254.1
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr1_+_156308245 | 0.13 |
ENST00000368253.2
ENST00000470342.1 ENST00000368254.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr1_+_156308403 | 0.13 |
ENST00000481479.1
ENST00000368252.1 ENST00000466306.1 ENST00000368251.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr12_-_9885702 | 0.12 |
ENST00000542530.1
|
CLECL1
|
C-type lectin-like 1 |
| chr10_+_24528108 | 0.12 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chr4_+_5527117 | 0.12 |
ENST00000505296.1
|
C4orf6
|
chromosome 4 open reading frame 6 |
| chr17_+_34087888 | 0.12 |
ENST00000586491.1
ENST00000588628.1 ENST00000285023.4 |
C17orf50
|
chromosome 17 open reading frame 50 |
| chr22_+_41253080 | 0.12 |
ENST00000541156.1
ENST00000414396.1 ENST00000357137.4 |
XPNPEP3
|
X-prolyl aminopeptidase (aminopeptidase P) 3, putative |
| chrX_+_15767971 | 0.12 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr11_+_193065 | 0.11 |
ENST00000342878.2
|
SCGB1C1
|
secretoglobin, family 1C, member 1 |
| chr1_-_205904950 | 0.11 |
ENST00000340781.4
|
SLC26A9
|
solute carrier family 26 (anion exchanger), member 9 |
| chr19_-_344786 | 0.11 |
ENST00000264819.4
|
MIER2
|
mesoderm induction early response 1, family member 2 |
| chr19_+_751122 | 0.11 |
ENST00000215582.6
|
MISP
|
mitotic spindle positioning |
| chr1_-_149459549 | 0.11 |
ENST00000369175.3
|
FAM72C
|
family with sequence similarity 72, member C |
| chr17_-_76573465 | 0.11 |
ENST00000585328.1
ENST00000389840.5 |
DNAH17
|
dynein, axonemal, heavy chain 17 |
| chr12_-_4754339 | 0.11 |
ENST00000228850.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr1_+_154947148 | 0.11 |
ENST00000368436.1
ENST00000308987.5 |
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr5_+_10250328 | 0.11 |
ENST00000515390.1
|
CCT5
|
chaperonin containing TCP1, subunit 5 (epsilon) |
| chr1_-_26633067 | 0.10 |
ENST00000421827.2
ENST00000374215.1 ENST00000374223.1 ENST00000357089.4 ENST00000535108.1 ENST00000314675.7 ENST00000436301.2 ENST00000423664.1 ENST00000374221.3 |
UBXN11
|
UBX domain protein 11 |
| chr17_+_74723031 | 0.10 |
ENST00000586200.1
|
METTL23
|
methyltransferase like 23 |
| chr4_+_41258786 | 0.10 |
ENST00000503431.1
ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr4_-_185269050 | 0.10 |
ENST00000511465.1
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr14_-_102553371 | 0.10 |
ENST00000553585.1
ENST00000216281.8 |
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr1_+_196743912 | 0.10 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chrX_+_108780062 | 0.10 |
ENST00000372106.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr20_+_10015678 | 0.10 |
ENST00000378392.1
ENST00000378380.3 |
ANKEF1
|
ankyrin repeat and EF-hand domain containing 1 |
| chr15_+_89921280 | 0.10 |
ENST00000560596.1
ENST00000558692.1 ENST00000538734.2 ENST00000559235.1 |
LINC00925
|
long intergenic non-protein coding RNA 925 |
| chrX_-_152939133 | 0.10 |
ENST00000370150.1
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr1_+_53925063 | 0.10 |
ENST00000371445.3
|
DMRTB1
|
DMRT-like family B with proline-rich C-terminal, 1 |
| chr7_-_100493744 | 0.10 |
ENST00000428317.1
ENST00000441605.1 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr1_+_154947126 | 0.10 |
ENST00000368439.1
|
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr6_+_31683117 | 0.09 |
ENST00000375825.3
ENST00000375824.1 |
LY6G6D
|
lymphocyte antigen 6 complex, locus G6D |
| chr21_+_45148735 | 0.09 |
ENST00000327574.4
|
PDXK
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr2_+_201171242 | 0.09 |
ENST00000360760.5
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr17_-_61850894 | 0.09 |
ENST00000403162.3
ENST00000582252.1 ENST00000225726.5 |
CCDC47
|
coiled-coil domain containing 47 |
| chr11_-_14913765 | 0.09 |
ENST00000334636.5
|
CYP2R1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chrX_+_150866779 | 0.09 |
ENST00000370353.3
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr5_+_118690466 | 0.09 |
ENST00000503646.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr2_-_208994548 | 0.09 |
ENST00000282141.3
|
CRYGC
|
crystallin, gamma C |
| chr19_-_52133588 | 0.09 |
ENST00000570106.2
|
SIGLEC5
|
sialic acid binding Ig-like lectin 5 |
| chr2_+_201171372 | 0.09 |
ENST00000409140.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr2_+_201170770 | 0.09 |
ENST00000409988.3
ENST00000409385.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr15_-_58357932 | 0.09 |
ENST00000347587.3
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr9_-_86322831 | 0.09 |
ENST00000257468.7
|
UBQLN1
|
ubiquilin 1 |
| chr4_-_159644507 | 0.08 |
ENST00000307720.3
|
PPID
|
peptidylprolyl isomerase D |
| chr5_-_13944652 | 0.08 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr7_-_139763521 | 0.08 |
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr1_+_196857144 | 0.08 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr19_-_17958771 | 0.08 |
ENST00000534444.1
|
JAK3
|
Janus kinase 3 |
| chr17_+_45401308 | 0.08 |
ENST00000331493.2
ENST00000519772.1 ENST00000517484.1 |
EFCAB13
|
EF-hand calcium binding domain 13 |
| chr20_+_55904815 | 0.08 |
ENST00000371263.3
ENST00000345868.4 ENST00000371260.4 ENST00000418127.1 |
SPO11
|
SPO11 meiotic protein covalently bound to DSB |
| chr2_+_85766280 | 0.08 |
ENST00000306434.3
|
MAT2A
|
methionine adenosyltransferase II, alpha |
| chr11_-_89956227 | 0.08 |
ENST00000457199.2
ENST00000530765.1 |
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1 |
| chr11_+_65154070 | 0.08 |
ENST00000317568.5
ENST00000531296.1 ENST00000533782.1 ENST00000355991.5 ENST00000416776.2 ENST00000526201.1 |
FRMD8
|
FERM domain containing 8 |
| chr4_+_76932326 | 0.07 |
ENST00000513353.1
ENST00000341029.5 |
ART3
|
ADP-ribosyltransferase 3 |
| chr6_+_44215603 | 0.07 |
ENST00000371554.1
|
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr1_-_9714644 | 0.07 |
ENST00000377320.3
|
C1orf200
|
chromosome 1 open reading frame 200 |
| chr3_+_138066539 | 0.07 |
ENST00000289104.4
|
MRAS
|
muscle RAS oncogene homolog |
| chr1_+_28586006 | 0.07 |
ENST00000253063.3
|
SESN2
|
sestrin 2 |
| chr2_-_62115659 | 0.07 |
ENST00000544185.1
|
CCT4
|
chaperonin containing TCP1, subunit 4 (delta) |
| chr18_+_57567180 | 0.07 |
ENST00000316660.6
ENST00000269518.9 |
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr11_-_89956461 | 0.07 |
ENST00000320585.6
|
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1 |
| chr9_+_34989638 | 0.07 |
ENST00000453597.3
ENST00000335998.3 ENST00000312316.5 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr14_-_23540826 | 0.07 |
ENST00000357481.2
|
ACIN1
|
apoptotic chromatin condensation inducer 1 |
| chr19_-_474880 | 0.07 |
ENST00000382696.3
ENST00000315489.4 |
ODF3L2
|
outer dense fiber of sperm tails 3-like 2 |
| chr14_-_23540747 | 0.07 |
ENST00000555566.1
ENST00000338631.6 ENST00000557515.1 ENST00000397341.3 |
ACIN1
|
apoptotic chromatin condensation inducer 1 |
| chrX_-_106243451 | 0.07 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr12_-_110883346 | 0.07 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr6_-_76782371 | 0.06 |
ENST00000369950.3
ENST00000369963.3 |
IMPG1
|
interphotoreceptor matrix proteoglycan 1 |
| chrX_-_20236970 | 0.06 |
ENST00000379548.4
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr6_-_32339649 | 0.06 |
ENST00000527965.1
ENST00000532023.1 ENST00000447241.2 ENST00000375007.4 ENST00000534588.1 |
C6orf10
|
chromosome 6 open reading frame 10 |
| chr7_+_130020180 | 0.06 |
ENST00000481342.1
ENST00000011292.3 ENST00000604896.1 |
CPA1
|
carboxypeptidase A1 (pancreatic) |
| chr12_-_9885888 | 0.06 |
ENST00000327839.3
|
CLECL1
|
C-type lectin-like 1 |
| chr2_-_62115725 | 0.06 |
ENST00000538252.1
ENST00000544079.1 ENST00000394440.3 |
CCT4
|
chaperonin containing TCP1, subunit 4 (delta) |
| chr17_-_74137374 | 0.06 |
ENST00000322957.6
|
FOXJ1
|
forkhead box J1 |
| chr21_+_31768348 | 0.06 |
ENST00000355459.2
|
KRTAP13-1
|
keratin associated protein 13-1 |
| chr1_+_161494036 | 0.06 |
ENST00000309758.4
|
HSPA6
|
heat shock 70kDa protein 6 (HSP70B') |
| chr6_-_32339671 | 0.06 |
ENST00000442822.2
ENST00000375015.4 ENST00000533191.1 |
C6orf10
|
chromosome 6 open reading frame 10 |
| chr20_+_19867150 | 0.06 |
ENST00000255006.6
|
RIN2
|
Ras and Rab interactor 2 |
| chr6_+_42531798 | 0.06 |
ENST00000372903.2
ENST00000372899.1 ENST00000372901.1 |
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr16_+_29911666 | 0.06 |
ENST00000563177.1
ENST00000483405.1 |
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr9_-_6015607 | 0.06 |
ENST00000259569.5
|
RANBP6
|
RAN binding protein 6 |
| chrX_-_102942961 | 0.05 |
ENST00000434230.1
ENST00000418819.1 ENST00000360458.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr5_+_147258266 | 0.05 |
ENST00000296694.4
|
SCGB3A2
|
secretoglobin, family 3A, member 2 |
| chr5_-_140053152 | 0.05 |
ENST00000542735.1
|
DND1
|
DND microRNA-mediated repression inhibitor 1 |
| chr7_-_151107767 | 0.05 |
ENST00000477459.1
|
WDR86
|
WD repeat domain 86 |
| chrX_-_102943022 | 0.05 |
ENST00000433176.2
|
MORF4L2
|
mortality factor 4 like 2 |
| chr4_+_5526883 | 0.05 |
ENST00000195455.2
|
C4orf6
|
chromosome 4 open reading frame 6 |
| chr12_-_57505121 | 0.05 |
ENST00000538913.2
ENST00000537215.2 ENST00000454075.3 ENST00000554825.1 ENST00000553275.1 ENST00000300134.3 |
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr3_+_136676851 | 0.05 |
ENST00000309741.5
|
IL20RB
|
interleukin 20 receptor beta |
| chr8_+_143781883 | 0.05 |
ENST00000522591.1
|
LY6K
|
lymphocyte antigen 6 complex, locus K |
| chr22_+_29664248 | 0.05 |
ENST00000406548.1
ENST00000437155.2 ENST00000415761.1 ENST00000331029.7 |
EWSR1
|
EWS RNA-binding protein 1 |
| chrX_-_100548045 | 0.04 |
ENST00000372907.3
ENST00000372905.2 |
TAF7L
|
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa |
| chr1_-_24469602 | 0.04 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1 |
| chr4_-_152147579 | 0.04 |
ENST00000304527.4
ENST00000455740.1 ENST00000424281.1 ENST00000409598.4 |
SH3D19
|
SH3 domain containing 19 |
| chr20_+_55108302 | 0.04 |
ENST00000371325.1
|
FAM209B
|
family with sequence similarity 209, member B |
| chr12_+_6644443 | 0.04 |
ENST00000396858.1
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr1_-_146054494 | 0.04 |
ENST00000401009.2
|
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr14_-_23388338 | 0.04 |
ENST00000555209.1
ENST00000554256.1 ENST00000557403.1 ENST00000557549.1 ENST00000555676.1 ENST00000557571.1 ENST00000557464.1 ENST00000554618.1 ENST00000556862.1 ENST00000555722.1 ENST00000346528.5 ENST00000542016.2 ENST00000399922.2 ENST00000557227.1 ENST00000359890.3 |
RBM23
|
RNA binding motif protein 23 |
| chr10_-_90611566 | 0.04 |
ENST00000371930.4
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr11_-_47870091 | 0.04 |
ENST00000526870.1
|
NUP160
|
nucleoporin 160kDa |
| chr11_+_63953691 | 0.04 |
ENST00000543847.1
|
STIP1
|
stress-induced-phosphoprotein 1 |
| chr22_-_24181174 | 0.04 |
ENST00000318109.7
ENST00000406855.3 ENST00000404056.1 ENST00000476077.1 |
DERL3
|
derlin 3 |
| chr20_+_36149602 | 0.04 |
ENST00000062104.2
ENST00000346199.2 |
NNAT
|
neuronatin |
| chr9_-_138591341 | 0.04 |
ENST00000298466.5
ENST00000425225.1 |
SOHLH1
|
spermatogenesis and oogenesis specific basic helix-loop-helix 1 |
| chr19_+_859425 | 0.04 |
ENST00000327726.6
|
CFD
|
complement factor D (adipsin) |
| chr1_-_6453399 | 0.04 |
ENST00000608083.1
|
ACOT7
|
acyl-CoA thioesterase 7 |
| chrX_-_152989798 | 0.04 |
ENST00000441714.1
ENST00000442093.1 ENST00000429550.1 ENST00000345046.6 |
BCAP31
|
B-cell receptor-associated protein 31 |
| chrX_-_152989531 | 0.04 |
ENST00000458587.2
ENST00000416815.1 |
BCAP31
|
B-cell receptor-associated protein 31 |
| chr1_+_154966058 | 0.04 |
ENST00000392487.1
|
LENEP
|
lens epithelial protein |
| chr7_+_142378566 | 0.04 |
ENST00000390398.3
|
TRBV25-1
|
T cell receptor beta variable 25-1 |
| chr2_+_201171064 | 0.04 |
ENST00000451764.2
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr11_-_47870019 | 0.04 |
ENST00000378460.2
|
NUP160
|
nucleoporin 160kDa |
| chr14_-_68141535 | 0.04 |
ENST00000554659.1
|
VTI1B
|
vesicle transport through interaction with t-SNAREs 1B |
| chr11_+_111782934 | 0.04 |
ENST00000304298.3
|
HSPB2
|
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr15_+_79165151 | 0.03 |
ENST00000331268.5
|
MORF4L1
|
mortality factor 4 like 1 |
| chr8_+_143781513 | 0.03 |
ENST00000292430.6
ENST00000561179.1 ENST00000518841.1 ENST00000519387.1 |
LY6K
|
lymphocyte antigen 6 complex, locus K |
| chr1_-_228613026 | 0.03 |
ENST00000366696.1
|
HIST3H3
|
histone cluster 3, H3 |
| chr1_-_242162375 | 0.03 |
ENST00000357246.3
|
MAP1LC3C
|
microtubule-associated protein 1 light chain 3 gamma |
| chr19_-_55672037 | 0.03 |
ENST00000588076.1
|
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr19_+_859654 | 0.03 |
ENST00000592860.1
|
CFD
|
complement factor D (adipsin) |
| chr11_-_108408895 | 0.03 |
ENST00000443411.1
ENST00000533052.1 |
EXPH5
|
exophilin 5 |
| chr17_-_54911250 | 0.03 |
ENST00000575658.1
ENST00000397861.2 |
C17orf67
|
chromosome 17 open reading frame 67 |
| chr1_+_67773044 | 0.03 |
ENST00000262345.1
ENST00000371000.1 |
IL12RB2
|
interleukin 12 receptor, beta 2 |
| chr15_+_45028753 | 0.03 |
ENST00000338264.4
|
TRIM69
|
tripartite motif containing 69 |
| chr9_-_86593238 | 0.03 |
ENST00000351839.3
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K |
| chr15_+_79165112 | 0.03 |
ENST00000426013.2
|
MORF4L1
|
mortality factor 4 like 1 |
| chr11_-_47869865 | 0.03 |
ENST00000530326.1
ENST00000532747.1 |
NUP160
|
nucleoporin 160kDa |
| chr20_-_23807358 | 0.03 |
ENST00000304725.2
|
CST2
|
cystatin SA |
| chr3_+_136676707 | 0.03 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr2_+_198365095 | 0.03 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr14_-_57277163 | 0.03 |
ENST00000555006.1
|
OTX2
|
orthodenticle homeobox 2 |
| chr20_+_5451845 | 0.02 |
ENST00000430097.2
|
RP5-828H9.1
|
RP5-828H9.1 |
| chr11_+_63953587 | 0.02 |
ENST00000305218.4
ENST00000538945.1 |
STIP1
|
stress-induced-phosphoprotein 1 |
| chr9_+_123970052 | 0.02 |
ENST00000373823.3
|
GSN
|
gelsolin |
| chr11_+_66512303 | 0.02 |
ENST00000532565.2
|
C11orf80
|
chromosome 11 open reading frame 80 |
| chr11_+_124492749 | 0.02 |
ENST00000531667.1
ENST00000441174.3 ENST00000375005.4 |
TBRG1
|
transforming growth factor beta regulator 1 |
| chr11_-_82997371 | 0.02 |
ENST00000525503.1
|
CCDC90B
|
coiled-coil domain containing 90B |
| chr16_-_23568651 | 0.02 |
ENST00000563232.1
ENST00000563459.1 ENST00000449606.1 |
EARS2
|
glutamyl-tRNA synthetase 2, mitochondrial |
| chr11_+_76092353 | 0.02 |
ENST00000530460.1
ENST00000321844.4 |
RP11-111M22.2
|
Homo sapiens putative uncharacterized protein FLJ37770-like (LOC100506127), mRNA. |
| chr8_+_62200509 | 0.02 |
ENST00000519846.1
ENST00000518592.1 ENST00000325897.4 |
CLVS1
|
clavesin 1 |
| chr1_+_27248203 | 0.02 |
ENST00000321265.5
|
NUDC
|
nudC nuclear distribution protein |
| chr8_-_52721975 | 0.02 |
ENST00000356297.4
ENST00000543296.1 |
PXDNL
|
peroxidasin homolog (Drosophila)-like |
| chr22_+_29168652 | 0.02 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr3_+_39509163 | 0.02 |
ENST00000436143.2
ENST00000441980.2 ENST00000311042.6 |
MOBP
|
myelin-associated oligodendrocyte basic protein |
| chr22_+_32871224 | 0.02 |
ENST00000452138.1
ENST00000382058.3 ENST00000397426.1 |
FBXO7
|
F-box protein 7 |
| chr12_+_110940005 | 0.02 |
ENST00000409246.1
ENST00000392672.4 ENST00000409300.1 ENST00000409425.1 |
RAD9B
|
RAD9 homolog B (S. pombe) |
| chr11_+_65769550 | 0.02 |
ENST00000312175.2
ENST00000445560.2 ENST00000530204.1 |
BANF1
|
barrier to autointegration factor 1 |
| chr11_+_65769946 | 0.02 |
ENST00000533166.1
|
BANF1
|
barrier to autointegration factor 1 |
| chr12_+_110940111 | 0.02 |
ENST00000409778.3
|
RAD9B
|
RAD9 homolog B (S. pombe) |
Network of associatons between targets according to the STRING database.
First level regulatory network of HSF4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 0.7 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 1.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.7 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.1 | 0.2 | GO:0045210 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
| 0.1 | 0.2 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.2 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.1 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.3 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.1 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.1 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 0.1 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) meiotic DNA double-strand break formation(GO:0042138) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:1904869 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.1 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.2 | GO:0070970 | interleukin-2 secretion(GO:0070970) |
| 0.0 | 0.0 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.2 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.1 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.2 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.8 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.2 | GO:0005858 | axonemal dynein complex(GO:0005858) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.2 | 0.7 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0008478 | pyridoxal kinase activity(GO:0008478) lithium ion binding(GO:0031403) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.3 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |