Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
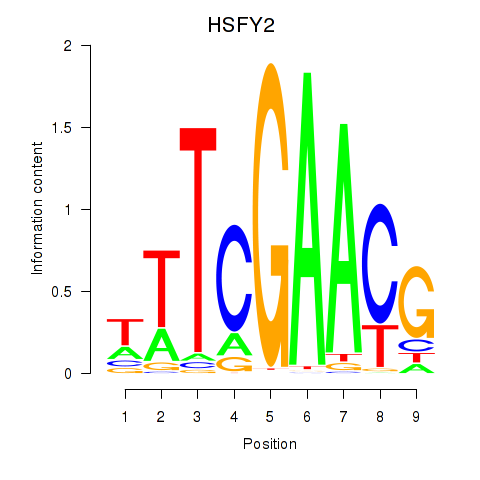
Results for HSFY2
Z-value: 0.56
Transcription factors associated with HSFY2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSFY2
|
ENSG00000169953.11 | heat shock transcription factor Y-linked 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSFY2 | hg19_v2_chrY_-_20935572_20935621 | 0.22 | 2.8e-01 | Click! |
Activity profile of HSFY2 motif
Sorted Z-values of HSFY2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_49860525 | 0.66 |
ENST00000435790.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr2_-_169769787 | 0.60 |
ENST00000451987.1
|
SPC25
|
SPC25, NDC80 kinetochore complex component |
| chr3_-_20227619 | 0.51 |
ENST00000425061.1
ENST00000443724.1 ENST00000421451.1 ENST00000452020.1 ENST00000417364.1 ENST00000306698.2 ENST00000419233.2 ENST00000263753.4 ENST00000383774.1 ENST00000437051.1 ENST00000412868.1 ENST00000429446.3 ENST00000442720.1 |
SGOL1
|
shugoshin-like 1 (S. pombe) |
| chr16_-_67969888 | 0.49 |
ENST00000574576.2
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr11_+_34654011 | 0.47 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr14_-_85996332 | 0.47 |
ENST00000380722.1
|
RP11-497E19.1
|
RP11-497E19.1 |
| chr5_-_135290705 | 0.45 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr14_+_21525981 | 0.43 |
ENST00000308227.2
|
RNASE8
|
ribonuclease, RNase A family, 8 |
| chr6_-_44281043 | 0.42 |
ENST00000244571.4
|
AARS2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr22_-_17073700 | 0.40 |
ENST00000359963.3
|
CCT8L2
|
chaperonin containing TCP1, subunit 8 (theta)-like 2 |
| chr4_-_100356291 | 0.38 |
ENST00000476959.1
ENST00000482593.1 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr2_+_1418154 | 0.38 |
ENST00000423320.1
ENST00000382198.1 |
TPO
|
thyroid peroxidase |
| chr10_+_114135004 | 0.38 |
ENST00000393081.1
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr13_-_41706864 | 0.37 |
ENST00000379485.1
ENST00000499385.2 |
KBTBD6
|
kelch repeat and BTB (POZ) domain containing 6 |
| chr10_+_114133773 | 0.37 |
ENST00000354655.4
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr7_+_138943265 | 0.35 |
ENST00000483726.1
|
UBN2
|
ubinuclein 2 |
| chr12_+_66217911 | 0.34 |
ENST00000403681.2
|
HMGA2
|
high mobility group AT-hook 2 |
| chr2_+_234959323 | 0.33 |
ENST00000373368.1
ENST00000168148.3 |
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr15_-_80263506 | 0.33 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr13_-_33112823 | 0.33 |
ENST00000504114.1
|
N4BP2L2
|
NEDD4 binding protein 2-like 2 |
| chr9_+_470288 | 0.31 |
ENST00000382303.1
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr12_+_113376157 | 0.31 |
ENST00000228928.7
|
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr5_+_42756903 | 0.31 |
ENST00000361970.5
ENST00000388827.4 |
CCDC152
|
coiled-coil domain containing 152 |
| chr11_+_5372738 | 0.29 |
ENST00000380219.1
|
OR51B6
|
olfactory receptor, family 51, subfamily B, member 6 |
| chr15_-_85197501 | 0.27 |
ENST00000434634.2
|
WDR73
|
WD repeat domain 73 |
| chr1_-_67142710 | 0.27 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr13_+_76378357 | 0.27 |
ENST00000489941.2
ENST00000525373.1 |
LMO7
|
LIM domain 7 |
| chr12_+_109785708 | 0.27 |
ENST00000310903.5
|
MYO1H
|
myosin IH |
| chr10_+_106113515 | 0.27 |
ENST00000369704.3
ENST00000312902.5 |
CCDC147
|
coiled-coil domain containing 147 |
| chr6_-_29343068 | 0.27 |
ENST00000396806.3
|
OR12D3
|
olfactory receptor, family 12, subfamily D, member 3 |
| chr2_+_74056066 | 0.26 |
ENST00000339566.3
ENST00000409707.1 ENST00000452725.1 ENST00000432295.2 ENST00000424659.1 ENST00000394073.1 |
STAMBP
|
STAM binding protein |
| chrX_+_38211777 | 0.26 |
ENST00000039007.4
|
OTC
|
ornithine carbamoyltransferase |
| chr9_-_25678856 | 0.26 |
ENST00000358022.3
|
TUSC1
|
tumor suppressor candidate 1 |
| chr9_-_99180597 | 0.26 |
ENST00000375256.4
|
ZNF367
|
zinc finger protein 367 |
| chrX_+_56259316 | 0.26 |
ENST00000468660.1
|
KLF8
|
Kruppel-like factor 8 |
| chr6_+_125524785 | 0.25 |
ENST00000392482.2
|
TPD52L1
|
tumor protein D52-like 1 |
| chr11_-_88796803 | 0.24 |
ENST00000418177.2
ENST00000455756.2 |
GRM5
|
glutamate receptor, metabotropic 5 |
| chr3_+_142838091 | 0.24 |
ENST00000309575.3
|
CHST2
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 |
| chr2_+_39005336 | 0.24 |
ENST00000409566.1
|
GEMIN6
|
gem (nuclear organelle) associated protein 6 |
| chr19_+_58341656 | 0.24 |
ENST00000442832.4
ENST00000594901.1 |
ZNF587B
|
zinc finger protein 587B |
| chr18_+_51795774 | 0.24 |
ENST00000579534.1
ENST00000406285.3 ENST00000577612.1 ENST00000579434.1 ENST00000583136.1 |
POLI
|
polymerase (DNA directed) iota |
| chr12_-_54691668 | 0.24 |
ENST00000553198.1
|
NFE2
|
nuclear factor, erythroid 2 |
| chr19_-_14952689 | 0.24 |
ENST00000248058.1
|
OR7A10
|
olfactory receptor, family 7, subfamily A, member 10 |
| chr15_+_71389281 | 0.23 |
ENST00000355327.3
|
THSD4
|
thrombospondin, type I, domain containing 4 |
| chr1_+_28052456 | 0.23 |
ENST00000373954.6
ENST00000419687.2 |
FAM76A
|
family with sequence similarity 76, member A |
| chr3_-_146187088 | 0.23 |
ENST00000497985.1
|
PLSCR2
|
phospholipid scramblase 2 |
| chr6_-_39902160 | 0.23 |
ENST00000340692.5
|
MOCS1
|
molybdenum cofactor synthesis 1 |
| chr4_+_118955500 | 0.23 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr12_-_132628847 | 0.23 |
ENST00000397333.3
|
DDX51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 |
| chr4_+_9172135 | 0.23 |
ENST00000512047.1
|
FAM90A26
|
family with sequence similarity 90, member A26 |
| chr14_+_96722539 | 0.23 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr1_-_39339777 | 0.22 |
ENST00000397572.2
|
MYCBP
|
MYC binding protein |
| chr3_+_173116225 | 0.22 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr20_+_814377 | 0.22 |
ENST00000304189.2
ENST00000381939.1 |
FAM110A
|
family with sequence similarity 110, member A |
| chr13_-_28024681 | 0.22 |
ENST00000381116.1
ENST00000381120.3 ENST00000431572.2 |
MTIF3
|
mitochondrial translational initiation factor 3 |
| chr1_+_1109272 | 0.22 |
ENST00000379290.1
ENST00000379289.1 |
TTLL10
|
tubulin tyrosine ligase-like family, member 10 |
| chr22_+_23241661 | 0.22 |
ENST00000390322.2
|
IGLJ2
|
immunoglobulin lambda joining 2 |
| chr19_-_4540486 | 0.22 |
ENST00000306390.6
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr6_+_127898312 | 0.22 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr11_-_506316 | 0.21 |
ENST00000532055.1
ENST00000531540.1 |
RNH1
|
ribonuclease/angiogenin inhibitor 1 |
| chr11_+_124543694 | 0.21 |
ENST00000227135.2
ENST00000532692.1 |
SPA17
|
sperm autoantigenic protein 17 |
| chr13_+_25875785 | 0.21 |
ENST00000381747.3
|
NUPL1
|
nucleoporin like 1 |
| chr10_-_101380121 | 0.21 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr11_+_111126707 | 0.21 |
ENST00000280325.4
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chr15_+_75074385 | 0.21 |
ENST00000220003.9
|
CSK
|
c-src tyrosine kinase |
| chr10_-_102046417 | 0.20 |
ENST00000370372.2
|
BLOC1S2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr3_+_155838337 | 0.20 |
ENST00000490337.1
ENST00000389636.5 |
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr17_+_55163075 | 0.20 |
ENST00000571629.1
ENST00000570423.1 ENST00000575186.1 ENST00000573085.1 ENST00000572814.1 |
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr2_-_55496344 | 0.20 |
ENST00000403721.1
ENST00000263629.4 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr11_+_17281900 | 0.20 |
ENST00000530527.1
|
NUCB2
|
nucleobindin 2 |
| chr1_+_111888890 | 0.20 |
ENST00000369738.4
|
PIFO
|
primary cilia formation |
| chr2_+_127413677 | 0.20 |
ENST00000356887.7
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr1_-_145075847 | 0.20 |
ENST00000530740.1
ENST00000369359.4 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr1_-_109203997 | 0.20 |
ENST00000370032.5
|
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr10_-_22292613 | 0.20 |
ENST00000376980.3
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr5_-_37371278 | 0.20 |
ENST00000231498.3
|
NUP155
|
nucleoporin 155kDa |
| chr17_-_74533963 | 0.20 |
ENST00000293230.5
|
CYGB
|
cytoglobin |
| chr10_-_95462265 | 0.20 |
ENST00000536233.1
ENST00000359204.4 ENST00000371430.2 ENST00000394100.2 |
FRA10AC1
|
fragile site, folic acid type, rare, fra(10)(q23.3) or fra(10)(q24.2) candidate 1 |
| chrX_+_46940254 | 0.19 |
ENST00000336169.3
|
RGN
|
regucalcin |
| chr3_-_52804872 | 0.19 |
ENST00000535191.1
ENST00000461689.1 ENST00000383721.4 ENST00000233027.5 |
NEK4
|
NIMA-related kinase 4 |
| chr1_-_117210290 | 0.19 |
ENST00000369483.1
ENST00000369486.3 |
IGSF3
|
immunoglobulin superfamily, member 3 |
| chr9_+_125273081 | 0.19 |
ENST00000335302.5
|
OR1J2
|
olfactory receptor, family 1, subfamily J, member 2 |
| chr7_-_32338917 | 0.19 |
ENST00000396193.1
|
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr3_+_122399444 | 0.19 |
ENST00000474629.2
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr10_-_102046098 | 0.19 |
ENST00000441611.1
|
BLOC1S2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr1_+_54569968 | 0.19 |
ENST00000391366.1
|
AL161915.1
|
Uncharacterized protein |
| chr13_+_37581115 | 0.19 |
ENST00000481013.1
|
EXOSC8
|
exosome component 8 |
| chr16_-_20367584 | 0.19 |
ENST00000570689.1
|
UMOD
|
uromodulin |
| chr7_-_102789629 | 0.19 |
ENST00000417955.1
ENST00000341533.4 ENST00000425379.1 |
NAPEPLD
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr1_-_150207017 | 0.19 |
ENST00000369119.3
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr6_-_155635583 | 0.18 |
ENST00000367166.4
|
TFB1M
|
transcription factor B1, mitochondrial |
| chr15_+_89182156 | 0.18 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr1_-_197115818 | 0.18 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr8_+_124084899 | 0.18 |
ENST00000287380.1
ENST00000309336.3 ENST00000519418.1 ENST00000327098.5 ENST00000522420.1 ENST00000521676.1 ENST00000378080.2 |
TBC1D31
|
TBC1 domain family, member 31 |
| chr3_-_72149553 | 0.18 |
ENST00000468646.2
ENST00000464271.1 |
LINC00877
|
long intergenic non-protein coding RNA 877 |
| chr16_+_11038345 | 0.18 |
ENST00000409790.1
|
CLEC16A
|
C-type lectin domain family 16, member A |
| chr16_+_31483374 | 0.18 |
ENST00000394863.3
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chr1_-_94344754 | 0.18 |
ENST00000436063.2
|
DNTTIP2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr9_+_96793076 | 0.17 |
ENST00000375360.3
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chr2_+_36923933 | 0.17 |
ENST00000497382.1
ENST00000404084.1 ENST00000379241.3 ENST00000401530.1 |
VIT
|
vitrin |
| chrX_-_153979315 | 0.17 |
ENST00000369575.3
ENST00000369568.4 ENST00000424127.2 |
GAB3
|
GRB2-associated binding protein 3 |
| chr6_+_30614779 | 0.17 |
ENST00000293604.6
ENST00000376473.5 |
C6orf136
|
chromosome 6 open reading frame 136 |
| chr16_-_5147743 | 0.17 |
ENST00000587133.1
ENST00000458008.4 ENST00000427587.4 |
FAM86A
|
family with sequence similarity 86, member A |
| chr22_-_45559540 | 0.17 |
ENST00000432502.1
|
CTA-217C2.1
|
CTA-217C2.1 |
| chr2_+_234959376 | 0.17 |
ENST00000425558.1
|
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr12_+_68042495 | 0.17 |
ENST00000344096.3
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr16_-_84220604 | 0.17 |
ENST00000567759.1
|
TAF1C
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa |
| chr19_+_51273721 | 0.17 |
ENST00000270590.4
|
GPR32
|
G protein-coupled receptor 32 |
| chr6_-_39902185 | 0.16 |
ENST00000373195.3
ENST00000308559.7 ENST00000373188.2 |
MOCS1
|
molybdenum cofactor synthesis 1 |
| chr19_+_44576296 | 0.16 |
ENST00000421176.3
|
ZNF284
|
zinc finger protein 284 |
| chr5_+_178368186 | 0.16 |
ENST00000320129.3
ENST00000519564.1 |
ZNF454
|
zinc finger protein 454 |
| chr6_+_132891461 | 0.16 |
ENST00000275198.1
|
TAAR6
|
trace amine associated receptor 6 |
| chr14_+_39583427 | 0.16 |
ENST00000308317.6
ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2
|
gem (nuclear organelle) associated protein 2 |
| chr20_-_62203808 | 0.16 |
ENST00000467148.1
|
HELZ2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr3_-_50336826 | 0.16 |
ENST00000443842.1
ENST00000354862.4 ENST00000443094.2 ENST00000415204.1 ENST00000336307.1 |
NAT6
HYAL3
|
N-acetyltransferase 6 (GCN5-related) hyaluronoglucosaminidase 3 |
| chr1_+_111889212 | 0.16 |
ENST00000369737.4
|
PIFO
|
primary cilia formation |
| chr11_-_62439012 | 0.16 |
ENST00000532208.1
ENST00000377954.2 ENST00000415855.2 ENST00000431002.2 ENST00000354588.3 |
C11orf48
|
chromosome 11 open reading frame 48 |
| chr1_+_234509413 | 0.16 |
ENST00000366613.1
ENST00000366612.1 |
COA6
|
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
| chr4_+_113558272 | 0.16 |
ENST00000509061.1
ENST00000508577.1 ENST00000513553.1 |
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr17_-_78428487 | 0.16 |
ENST00000562672.2
|
CTD-2526A2.2
|
CTD-2526A2.2 |
| chr9_+_99212403 | 0.16 |
ENST00000375251.3
ENST00000375249.4 |
HABP4
|
hyaluronan binding protein 4 |
| chr1_+_62417957 | 0.16 |
ENST00000307297.7
ENST00000543708.1 |
INADL
|
InaD-like (Drosophila) |
| chr3_+_184053703 | 0.16 |
ENST00000450976.1
ENST00000418281.1 ENST00000340957.5 ENST00000433578.1 |
FAM131A
|
family with sequence similarity 131, member A |
| chr1_+_55181490 | 0.16 |
ENST00000371281.3
|
TTC4
|
tetratricopeptide repeat domain 4 |
| chrX_-_33146477 | 0.15 |
ENST00000378677.2
|
DMD
|
dystrophin |
| chr2_+_65216462 | 0.15 |
ENST00000234256.3
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr12_+_14572070 | 0.15 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr22_-_45559642 | 0.15 |
ENST00000426282.2
|
CTA-217C2.1
|
CTA-217C2.1 |
| chr6_+_110501621 | 0.15 |
ENST00000368930.1
ENST00000307731.1 |
CDC40
|
cell division cycle 40 |
| chr12_+_7053228 | 0.15 |
ENST00000540506.2
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr17_-_46682321 | 0.15 |
ENST00000225648.3
ENST00000484302.2 |
HOXB6
|
homeobox B6 |
| chr2_+_220492287 | 0.15 |
ENST00000273063.6
ENST00000373762.3 |
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr4_+_76871883 | 0.15 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr14_+_105267250 | 0.15 |
ENST00000342537.7
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr19_+_58193337 | 0.15 |
ENST00000601064.1
ENST00000282296.5 ENST00000356715.4 |
ZNF551
|
zinc finger protein 551 |
| chr9_+_131037623 | 0.15 |
ENST00000495313.1
ENST00000372898.2 |
SWI5
|
SWI5 recombination repair homolog (yeast) |
| chr12_+_107168418 | 0.15 |
ENST00000392839.2
ENST00000548914.1 ENST00000355478.2 ENST00000552619.1 ENST00000549643.1 |
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr4_+_119606523 | 0.15 |
ENST00000388822.5
ENST00000506780.1 ENST00000508801.1 |
METTL14
|
methyltransferase like 14 |
| chr19_+_50529212 | 0.15 |
ENST00000270617.3
ENST00000445728.3 ENST00000601364.1 |
ZNF473
|
zinc finger protein 473 |
| chr19_+_33182823 | 0.15 |
ENST00000397061.3
|
NUDT19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
| chr5_+_149109825 | 0.15 |
ENST00000360453.4
ENST00000394320.3 ENST00000309241.5 |
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr1_-_222763240 | 0.15 |
ENST00000352967.4
ENST00000391882.1 ENST00000543857.1 |
TAF1A
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa |
| chr6_+_163148161 | 0.14 |
ENST00000337019.3
ENST00000366889.2 |
PACRG
|
PARK2 co-regulated |
| chr2_+_220492116 | 0.14 |
ENST00000373760.2
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr15_+_84115868 | 0.14 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr7_-_91509986 | 0.14 |
ENST00000456229.1
ENST00000442961.1 ENST00000406735.2 ENST00000419292.1 ENST00000351870.3 |
MTERF
|
mitochondrial transcription termination factor |
| chr9_+_95820966 | 0.14 |
ENST00000375472.3
ENST00000465709.1 |
SUSD3
|
sushi domain containing 3 |
| chr9_-_6015607 | 0.14 |
ENST00000259569.5
|
RANBP6
|
RAN binding protein 6 |
| chr21_-_37838739 | 0.14 |
ENST00000399139.1
|
CLDN14
|
claudin 14 |
| chr15_+_84116106 | 0.14 |
ENST00000535412.1
ENST00000324537.5 |
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr10_-_43133950 | 0.14 |
ENST00000359467.3
|
ZNF33B
|
zinc finger protein 33B |
| chr10_-_22292675 | 0.14 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr8_+_22224811 | 0.14 |
ENST00000381237.1
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr2_+_89986318 | 0.14 |
ENST00000491977.1
|
IGKV2D-29
|
immunoglobulin kappa variable 2D-29 |
| chr4_-_104119528 | 0.14 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr21_-_40720974 | 0.14 |
ENST00000380748.1
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr1_+_214776516 | 0.14 |
ENST00000366955.3
|
CENPF
|
centromere protein F, 350/400kDa |
| chr15_+_75074410 | 0.14 |
ENST00000439220.2
|
CSK
|
c-src tyrosine kinase |
| chr11_+_62439126 | 0.14 |
ENST00000377953.3
|
C11orf83
|
chromosome 11 open reading frame 83 |
| chr1_-_89458415 | 0.13 |
ENST00000321792.5
ENST00000370491.3 |
RBMXL1
CCBL2
|
RNA binding motif protein, X-linked-like 1 cysteine conjugate-beta lyase 2 |
| chr12_-_48419165 | 0.13 |
ENST00000547602.1
|
RP1-228P16.5
|
Uncharacterized protein |
| chr15_-_41624685 | 0.13 |
ENST00000560640.1
ENST00000220514.3 |
OIP5
|
Opa interacting protein 5 |
| chr5_-_19988339 | 0.13 |
ENST00000382275.1
|
CDH18
|
cadherin 18, type 2 |
| chr15_-_57025759 | 0.13 |
ENST00000267807.7
|
ZNF280D
|
zinc finger protein 280D |
| chr12_-_50419177 | 0.13 |
ENST00000454520.2
ENST00000546595.1 ENST00000548824.1 ENST00000549777.1 ENST00000546723.1 ENST00000427314.2 ENST00000552157.1 ENST00000552310.1 ENST00000548644.1 ENST00000312377.5 ENST00000546786.1 ENST00000550149.1 ENST00000546764.1 ENST00000552004.1 ENST00000548320.1 ENST00000547905.1 ENST00000550651.1 ENST00000551145.1 ENST00000434422.1 ENST00000552921.1 |
RACGAP1
|
Rac GTPase activating protein 1 |
| chr5_+_122181184 | 0.13 |
ENST00000513881.1
|
SNX24
|
sorting nexin 24 |
| chr12_-_42877408 | 0.13 |
ENST00000552240.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr2_+_219110149 | 0.13 |
ENST00000456575.1
|
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr2_+_209131059 | 0.13 |
ENST00000422495.1
ENST00000452564.1 |
PIKFYVE
|
phosphoinositide kinase, FYVE finger containing |
| chr5_+_122181279 | 0.13 |
ENST00000395451.4
ENST00000506996.1 |
SNX24
|
sorting nexin 24 |
| chr1_+_93646238 | 0.13 |
ENST00000448243.1
ENST00000370276.1 |
CCDC18
|
coiled-coil domain containing 18 |
| chr3_+_23847394 | 0.13 |
ENST00000306627.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr16_+_50775971 | 0.13 |
ENST00000311559.9
ENST00000564326.1 ENST00000566206.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr3_+_196669494 | 0.13 |
ENST00000602845.1
|
NCBP2-AS2
|
NCBP2 antisense RNA 2 (head to head) |
| chr10_-_129691195 | 0.13 |
ENST00000368671.3
|
CLRN3
|
clarin 3 |
| chr9_-_74675521 | 0.13 |
ENST00000377024.3
|
C9orf57
|
chromosome 9 open reading frame 57 |
| chr17_-_38574169 | 0.13 |
ENST00000423485.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr2_+_36923830 | 0.13 |
ENST00000379242.3
ENST00000389975.3 |
VIT
|
vitrin |
| chr15_+_39542867 | 0.13 |
ENST00000318578.3
ENST00000561223.1 |
C15orf54
|
chromosome 15 open reading frame 54 |
| chr16_-_84220633 | 0.13 |
ENST00000566732.1
ENST00000561955.1 ENST00000564454.1 ENST00000341690.6 ENST00000541676.1 ENST00000570117.1 ENST00000564345.1 |
TAF1C
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa |
| chr13_-_24463530 | 0.13 |
ENST00000382172.3
|
MIPEP
|
mitochondrial intermediate peptidase |
| chr10_-_71930222 | 0.13 |
ENST00000458634.2
ENST00000373239.2 ENST00000373242.2 ENST00000373241.4 |
SAR1A
|
SAR1 homolog A (S. cerevisiae) |
| chr15_+_89181974 | 0.13 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr19_-_52489923 | 0.12 |
ENST00000593596.1
ENST00000243644.4 ENST00000594929.1 ENST00000601430.1 |
ZNF350
|
zinc finger protein 350 |
| chr3_+_62936098 | 0.12 |
ENST00000475886.1
ENST00000465684.1 ENST00000465262.1 ENST00000468072.1 |
LINC00698
|
long intergenic non-protein coding RNA 698 |
| chr12_+_66218212 | 0.12 |
ENST00000393578.3
ENST00000425208.2 ENST00000536545.1 ENST00000354636.3 |
HMGA2
|
high mobility group AT-hook 2 |
| chrX_+_100353153 | 0.12 |
ENST00000423383.1
ENST00000218507.5 ENST00000403304.2 ENST00000435570.1 |
CENPI
|
centromere protein I |
| chr15_+_45694523 | 0.12 |
ENST00000305560.6
|
SPATA5L1
|
spermatogenesis associated 5-like 1 |
| chr10_-_115614127 | 0.12 |
ENST00000369305.1
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr3_+_3168600 | 0.12 |
ENST00000251607.6
ENST00000339437.6 ENST00000280591.6 ENST00000420393.1 |
TRNT1
|
tRNA nucleotidyl transferase, CCA-adding, 1 |
| chr7_-_138363824 | 0.12 |
ENST00000419765.3
|
SVOPL
|
SVOP-like |
| chr17_+_30469579 | 0.12 |
ENST00000354266.3
ENST00000581094.1 ENST00000394692.2 |
RHOT1
|
ras homolog family member T1 |
| chr16_+_20817839 | 0.12 |
ENST00000348433.6
ENST00000568501.1 ENST00000566276.1 |
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr1_-_53163992 | 0.12 |
ENST00000371538.3
|
SELRC1
|
cytochrome c oxidase assembly factor 7 |
| chr18_-_48351743 | 0.12 |
ENST00000588444.1
ENST00000256425.2 ENST00000428869.2 |
MRO
|
maestro |
| chr9_+_37485932 | 0.12 |
ENST00000377798.4
ENST00000442009.2 |
POLR1E
|
polymerase (RNA) I polypeptide E, 53kDa |
| chr15_+_69365265 | 0.12 |
ENST00000415504.1
|
LINC00277
|
long intergenic non-protein coding RNA 277 |
| chr8_+_144120648 | 0.12 |
ENST00000395172.1
|
C8orf31
|
chromosome 8 open reading frame 31 |
| chr19_+_44716678 | 0.12 |
ENST00000586228.1
ENST00000588219.1 ENST00000313040.7 ENST00000589707.1 ENST00000588394.1 ENST00000589005.1 |
ZNF227
|
zinc finger protein 227 |
| chr9_-_35815013 | 0.12 |
ENST00000259667.5
|
HINT2
|
histidine triad nucleotide binding protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HSFY2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.4 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.5 | GO:0003131 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.1 | 0.4 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.1 | 0.3 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.5 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.3 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.1 | 0.3 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 0.3 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.1 | 0.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.2 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 | 0.2 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.1 | 0.3 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 0.2 | GO:0001189 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.4 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.2 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.2 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.2 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.3 | GO:1903998 | response to isolation stress(GO:0035900) regulation of eating behavior(GO:1903998) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.3 | GO:0042997 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.5 | GO:0051189 | molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0071029 | nuclear ncRNA surveillance(GO:0071029) nuclear polyadenylation-dependent rRNA catabolic process(GO:0071035) nuclear polyadenylation-dependent ncRNA catabolic process(GO:0071046) |
| 0.0 | 0.1 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.3 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.2 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.1 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.1 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.9 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.2 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.3 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.0 | 0.4 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.0 | 0.1 | GO:1905154 | negative regulation of synaptic vesicle recycling(GO:1903422) negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 | 0.2 | GO:0015811 | L-alanine transport(GO:0015808) L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:0051586 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.1 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.2 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.3 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.1 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:2000232 | regulation of rRNA processing(GO:2000232) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0071600 | otic vesicle morphogenesis(GO:0071600) |
| 0.0 | 0.4 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.0 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.2 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.5 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.0 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.0 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.0 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.1 | 0.6 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.2 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.1 | 0.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.4 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.5 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.3 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.5 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 0.5 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.1 | 0.3 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.4 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.1 | 0.4 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 0.2 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.2 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.2 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.0 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.0 | 0.3 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.1 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.0 | 0.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0043813 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.2 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) L-cystine transmembrane transporter activity(GO:0015184) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.1 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.4 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.7 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.2 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.0 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |