Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
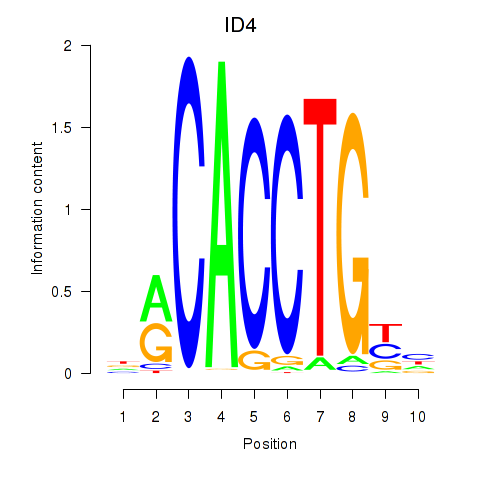
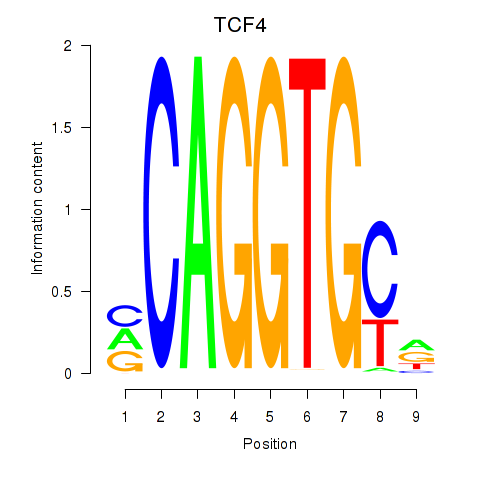
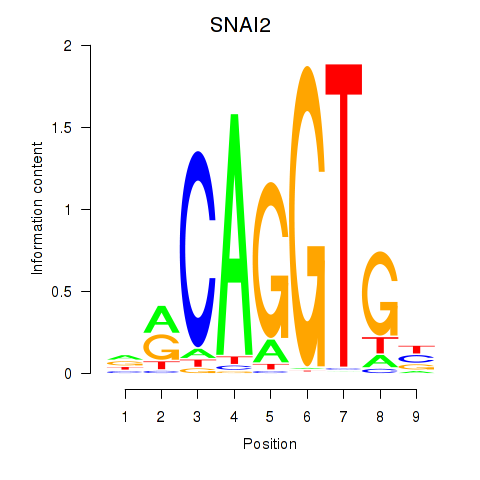
Results for ID4_TCF4_SNAI2
Z-value: 0.86
Transcription factors associated with ID4_TCF4_SNAI2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ID4
|
ENSG00000172201.6 | inhibitor of DNA binding 4, HLH protein |
|
TCF4
|
ENSG00000196628.9 | transcription factor 4 |
|
SNAI2
|
ENSG00000019549.4 | snail family transcriptional repressor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ID4 | hg19_v2_chr6_+_19837592_19837621 | 0.19 | 3.7e-01 | Click! |
| TCF4 | hg19_v2_chr18_-_52989217_52989233, hg19_v2_chr18_-_53070913_53070945 | 0.14 | 5.0e-01 | Click! |
| SNAI2 | hg19_v2_chr8_-_49834299_49834446, hg19_v2_chr8_-_49833978_49833996 | -0.13 | 5.4e-01 | Click! |
Activity profile of ID4_TCF4_SNAI2 motif
Sorted Z-values of ID4_TCF4_SNAI2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_39382900 | 2.11 |
ENST00000377721.3
ENST00000455970.2 |
KRTAP9-2
|
keratin associated protein 9-2 |
| chr4_+_156588350 | 1.40 |
ENST00000296518.7
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr4_+_156588115 | 1.39 |
ENST00000455639.2
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr4_+_156588249 | 1.38 |
ENST00000393832.3
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr19_-_7939319 | 1.30 |
ENST00000539422.1
|
CTD-3193O13.9
|
Protein FLJ22184 |
| chr5_-_94620239 | 1.24 |
ENST00000515393.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr17_+_39394250 | 1.24 |
ENST00000254072.6
|
KRTAP9-8
|
keratin associated protein 9-8 |
| chr4_+_156588806 | 1.21 |
ENST00000513574.1
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr19_+_6464502 | 1.21 |
ENST00000308243.7
|
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr4_+_156587979 | 1.17 |
ENST00000511507.1
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chrX_-_13956497 | 1.08 |
ENST00000398361.3
|
GPM6B
|
glycoprotein M6B |
| chr2_-_217559517 | 1.00 |
ENST00000449583.1
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr2_+_219745020 | 0.96 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr2_+_173600514 | 0.70 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr2_+_173600565 | 0.70 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr19_-_47137942 | 0.63 |
ENST00000300873.4
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr19_-_42721819 | 0.61 |
ENST00000336034.4
ENST00000598200.1 ENST00000598727.1 ENST00000596251.1 |
DEDD2
|
death effector domain containing 2 |
| chr11_+_45944190 | 0.61 |
ENST00000401752.1
ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr11_+_34663913 | 0.60 |
ENST00000532302.1
|
EHF
|
ets homologous factor |
| chr16_+_25228242 | 0.60 |
ENST00000219660.5
|
AQP8
|
aquaporin 8 |
| chr17_+_39405939 | 0.59 |
ENST00000334109.2
|
KRTAP9-4
|
keratin associated protein 9-4 |
| chr14_+_50999744 | 0.56 |
ENST00000441560.2
|
ATL1
|
atlastin GTPase 1 |
| chr1_+_214163033 | 0.56 |
ENST00000607425.1
|
PROX1
|
prospero homeobox 1 |
| chrX_-_117107680 | 0.56 |
ENST00000447671.2
ENST00000262820.3 |
KLHL13
|
kelch-like family member 13 |
| chrX_-_117107542 | 0.55 |
ENST00000371878.1
|
KLHL13
|
kelch-like family member 13 |
| chr14_-_107283278 | 0.55 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr14_-_69263043 | 0.53 |
ENST00000408913.2
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr14_+_105190514 | 0.53 |
ENST00000330877.2
|
ADSSL1
|
adenylosuccinate synthase like 1 |
| chr7_-_150675372 | 0.53 |
ENST00000262186.5
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr17_-_16256718 | 0.51 |
ENST00000476243.1
ENST00000299736.4 |
CENPV
|
centromere protein V |
| chr8_+_66955648 | 0.51 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr15_+_85427903 | 0.51 |
ENST00000286749.3
ENST00000394573.1 ENST00000537703.1 |
SLC28A1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr9_-_75567962 | 0.50 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr2_+_173600671 | 0.48 |
ENST00000409036.1
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr2_-_136873735 | 0.47 |
ENST00000409817.1
|
CXCR4
|
chemokine (C-X-C motif) receptor 4 |
| chr14_-_92414055 | 0.47 |
ENST00000342058.4
|
FBLN5
|
fibulin 5 |
| chr16_-_1968231 | 0.47 |
ENST00000443547.1
ENST00000293937.3 ENST00000454677.2 |
HS3ST6
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 6 |
| chr2_+_177053307 | 0.47 |
ENST00000331462.4
|
HOXD1
|
homeobox D1 |
| chr1_-_238054094 | 0.47 |
ENST00000366570.4
|
ZP4
|
zona pellucida glycoprotein 4 |
| chr2_-_24583583 | 0.46 |
ENST00000355123.4
|
ITSN2
|
intersectin 2 |
| chr1_+_2005425 | 0.46 |
ENST00000461106.2
|
PRKCZ
|
protein kinase C, zeta |
| chr7_-_142232071 | 0.45 |
ENST00000390364.3
|
TRBV10-1
|
T cell receptor beta variable 10-1(gene/pseudogene) |
| chr19_+_6464243 | 0.44 |
ENST00000600229.1
ENST00000356762.3 |
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr17_-_46035187 | 0.44 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr5_+_115298165 | 0.44 |
ENST00000357872.4
|
AQPEP
|
Aminopeptidase Q |
| chr19_+_54058073 | 0.43 |
ENST00000505949.1
ENST00000513265.1 |
ZNF331
|
zinc finger protein 331 |
| chr6_-_31080336 | 0.43 |
ENST00000259870.3
|
C6orf15
|
chromosome 6 open reading frame 15 |
| chr14_+_24779376 | 0.42 |
ENST00000530080.1
|
LTB4R2
|
leukotriene B4 receptor 2 |
| chr11_+_34654011 | 0.41 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr1_-_209979465 | 0.41 |
ENST00000542854.1
|
IRF6
|
interferon regulatory factor 6 |
| chrX_+_150863596 | 0.41 |
ENST00000448726.1
ENST00000538575.1 |
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr5_-_66492562 | 0.41 |
ENST00000256447.4
|
CD180
|
CD180 molecule |
| chr1_+_60280458 | 0.41 |
ENST00000455990.1
ENST00000371208.3 |
HOOK1
|
hook microtubule-tethering protein 1 |
| chr12_+_108908962 | 0.40 |
ENST00000552695.1
ENST00000552758.1 ENST00000361549.2 |
FICD
|
FIC domain containing |
| chr6_-_45983549 | 0.39 |
ENST00000544153.1
|
CLIC5
|
chloride intracellular channel 5 |
| chr17_-_53809473 | 0.39 |
ENST00000575734.1
|
TMEM100
|
transmembrane protein 100 |
| chr14_-_20929624 | 0.38 |
ENST00000398020.4
ENST00000250489.4 |
TMEM55B
|
transmembrane protein 55B |
| chr14_+_75746340 | 0.38 |
ENST00000555686.1
ENST00000555672.1 |
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr1_-_183387723 | 0.38 |
ENST00000287713.6
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr2_-_190445499 | 0.38 |
ENST00000261024.2
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr6_-_31697255 | 0.38 |
ENST00000436437.1
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr8_-_81083890 | 0.38 |
ENST00000518937.1
|
TPD52
|
tumor protein D52 |
| chr19_+_38755203 | 0.38 |
ENST00000587090.1
ENST00000454580.3 |
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr19_+_38755042 | 0.38 |
ENST00000301244.7
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr11_+_64879317 | 0.37 |
ENST00000526809.1
ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2
|
transmembrane 7 superfamily member 2 |
| chrX_-_152939252 | 0.37 |
ENST00000340888.3
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr7_-_8302298 | 0.37 |
ENST00000446305.1
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr7_-_73184588 | 0.37 |
ENST00000395145.2
|
CLDN3
|
claudin 3 |
| chr1_+_118148556 | 0.37 |
ENST00000369448.3
|
FAM46C
|
family with sequence similarity 46, member C |
| chr14_+_72399833 | 0.36 |
ENST00000553530.1
ENST00000556437.1 |
RGS6
|
regulator of G-protein signaling 6 |
| chr11_+_86667117 | 0.36 |
ENST00000531827.1
|
RP11-736K20.6
|
RP11-736K20.6 |
| chr16_-_3086927 | 0.35 |
ENST00000572449.1
|
CCDC64B
|
coiled-coil domain containing 64B |
| chr1_-_209979375 | 0.35 |
ENST00000367021.3
|
IRF6
|
interferon regulatory factor 6 |
| chr14_-_92413353 | 0.35 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr3_+_9975497 | 0.35 |
ENST00000397170.3
ENST00000383811.3 ENST00000452070.1 ENST00000326434.5 |
CRELD1
|
cysteine-rich with EGF-like domains 1 |
| chr16_+_4845379 | 0.35 |
ENST00000588606.1
ENST00000586005.1 |
SMIM22
|
small integral membrane protein 22 |
| chr2_+_205410516 | 0.35 |
ENST00000406610.2
ENST00000462231.1 |
PARD3B
|
par-3 family cell polarity regulator beta |
| chr22_+_19467261 | 0.35 |
ENST00000455750.1
ENST00000437685.2 ENST00000263201.1 ENST00000404724.3 |
CDC45
|
cell division cycle 45 |
| chr14_-_106967788 | 0.34 |
ENST00000390622.2
|
IGHV1-46
|
immunoglobulin heavy variable 1-46 |
| chr4_-_6202247 | 0.34 |
ENST00000409021.3
ENST00000409371.3 |
JAKMIP1
|
janus kinase and microtubule interacting protein 1 |
| chr16_+_68679193 | 0.34 |
ENST00000581171.1
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental) |
| chr1_+_177140633 | 0.34 |
ENST00000361539.4
|
BRINP2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chrX_-_152939133 | 0.34 |
ENST00000370150.1
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr22_-_36018569 | 0.34 |
ENST00000419229.1
ENST00000406324.1 |
MB
|
myoglobin |
| chr17_-_46507567 | 0.34 |
ENST00000584924.1
|
SKAP1
|
src kinase associated phosphoprotein 1 |
| chr17_-_78450398 | 0.34 |
ENST00000306773.4
|
NPTX1
|
neuronal pentraxin I |
| chr4_+_55524085 | 0.34 |
ENST00000412167.2
ENST00000288135.5 |
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
| chr21_-_46131470 | 0.34 |
ENST00000323084.4
|
TSPEAR
|
thrombospondin-type laminin G domain and EAR repeats |
| chr15_-_23891175 | 0.33 |
ENST00000532292.1
|
MAGEL2
|
MAGE-like 2 |
| chr7_-_144100786 | 0.33 |
ENST00000223140.5
|
NOBOX
|
NOBOX oogenesis homeobox |
| chr8_+_22132847 | 0.33 |
ENST00000521356.1
|
PIWIL2
|
piwi-like RNA-mediated gene silencing 2 |
| chrX_+_129473859 | 0.33 |
ENST00000424447.1
|
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr15_+_69591235 | 0.33 |
ENST00000395407.2
ENST00000558684.1 |
PAQR5
|
progestin and adipoQ receptor family member V |
| chr17_+_48912744 | 0.33 |
ENST00000311378.4
|
WFIKKN2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr11_+_34664014 | 0.32 |
ENST00000527935.1
|
EHF
|
ets homologous factor |
| chr1_-_1149506 | 0.32 |
ENST00000379236.3
|
TNFRSF4
|
tumor necrosis factor receptor superfamily, member 4 |
| chr8_-_141467818 | 0.32 |
ENST00000389327.3
ENST00000438773.2 |
TRAPPC9
|
trafficking protein particle complex 9 |
| chr8_+_98881268 | 0.32 |
ENST00000254898.5
ENST00000524308.1 ENST00000522025.2 |
MATN2
|
matrilin 2 |
| chr14_+_72399114 | 0.32 |
ENST00000553525.1
ENST00000555571.1 |
RGS6
|
regulator of G-protein signaling 6 |
| chr6_-_39290316 | 0.32 |
ENST00000425054.2
ENST00000373227.4 ENST00000373229.5 ENST00000437525.2 |
KCNK16
|
potassium channel, subfamily K, member 16 |
| chr4_+_1003742 | 0.32 |
ENST00000398484.2
|
FGFRL1
|
fibroblast growth factor receptor-like 1 |
| chr14_-_67981916 | 0.31 |
ENST00000357461.2
|
TMEM229B
|
transmembrane protein 229B |
| chr19_+_37342547 | 0.31 |
ENST00000331800.4
ENST00000586646.1 |
ZNF345
|
zinc finger protein 345 |
| chr16_+_23194033 | 0.31 |
ENST00000300061.2
|
SCNN1G
|
sodium channel, non-voltage-gated 1, gamma subunit |
| chr4_-_16085314 | 0.31 |
ENST00000510224.1
|
PROM1
|
prominin 1 |
| chr3_+_50712672 | 0.31 |
ENST00000266037.9
|
DOCK3
|
dedicator of cytokinesis 3 |
| chr19_-_3028354 | 0.31 |
ENST00000586422.1
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr6_+_22569784 | 0.31 |
ENST00000510882.2
|
HDGFL1
|
hepatoma derived growth factor-like 1 |
| chr4_+_142557771 | 0.31 |
ENST00000514653.1
|
IL15
|
interleukin 15 |
| chr19_+_37569314 | 0.30 |
ENST00000304239.7
ENST00000589245.1 ENST00000337995.3 |
ZNF420
|
zinc finger protein 420 |
| chr12_-_95941987 | 0.30 |
ENST00000537435.2
|
USP44
|
ubiquitin specific peptidase 44 |
| chr1_-_19186176 | 0.30 |
ENST00000375371.3
|
TAS1R2
|
taste receptor, type 1, member 2 |
| chr16_+_70680439 | 0.30 |
ENST00000288098.2
|
IL34
|
interleukin 34 |
| chr10_-_49701686 | 0.30 |
ENST00000417247.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr12_-_71551868 | 0.30 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr21_+_37507210 | 0.30 |
ENST00000290354.5
|
CBR3
|
carbonyl reductase 3 |
| chr12_-_71551652 | 0.30 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr1_-_115880852 | 0.30 |
ENST00000369512.2
|
NGF
|
nerve growth factor (beta polypeptide) |
| chr21_-_45079341 | 0.30 |
ENST00000443485.1
ENST00000291560.2 |
HSF2BP
|
heat shock transcription factor 2 binding protein |
| chr10_-_21463116 | 0.30 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chr19_+_11909329 | 0.30 |
ENST00000323169.5
ENST00000450087.1 |
ZNF491
|
zinc finger protein 491 |
| chr22_-_47882857 | 0.30 |
ENST00000405369.3
|
LL22NC03-75H12.2
|
Novel protein; Uncharacterized protein |
| chr3_+_32147997 | 0.30 |
ENST00000282541.5
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr11_+_1855645 | 0.29 |
ENST00000381968.3
ENST00000381978.3 |
SYT8
|
synaptotagmin VIII |
| chr19_-_51220176 | 0.29 |
ENST00000359082.3
ENST00000293441.1 |
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr8_-_72268889 | 0.29 |
ENST00000388742.4
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr16_-_11730213 | 0.29 |
ENST00000576334.1
ENST00000574848.1 |
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr6_+_150464155 | 0.28 |
ENST00000361131.4
|
PPP1R14C
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr7_+_5919458 | 0.28 |
ENST00000416608.1
|
OCM
|
oncomodulin |
| chr14_+_75746781 | 0.28 |
ENST00000555347.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr4_-_16085340 | 0.28 |
ENST00000508167.1
|
PROM1
|
prominin 1 |
| chr8_-_80993010 | 0.28 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr3_-_52479043 | 0.28 |
ENST00000231721.2
ENST00000475739.1 |
SEMA3G
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G |
| chr2_-_24583168 | 0.28 |
ENST00000361999.3
|
ITSN2
|
intersectin 2 |
| chr3_-_156534754 | 0.28 |
ENST00000472943.1
ENST00000473352.1 |
LINC00886
|
long intergenic non-protein coding RNA 886 |
| chr19_-_10420459 | 0.28 |
ENST00000403352.1
ENST00000403903.3 |
ZGLP1
|
zinc finger, GATA-like protein 1 |
| chr10_-_116286656 | 0.28 |
ENST00000428430.1
ENST00000369266.3 ENST00000392952.3 |
ABLIM1
|
actin binding LIM protein 1 |
| chr7_-_148580563 | 0.28 |
ENST00000476773.1
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr3_-_164875850 | 0.28 |
ENST00000472120.1
|
RP11-747D18.1
|
RP11-747D18.1 |
| chr4_+_156587853 | 0.28 |
ENST00000506455.1
ENST00000511108.1 |
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr6_+_132891461 | 0.27 |
ENST00000275198.1
|
TAAR6
|
trace amine associated receptor 6 |
| chr7_+_30960915 | 0.27 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr2_+_233527443 | 0.27 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr8_-_93029520 | 0.27 |
ENST00000521553.1
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr8_-_72268968 | 0.27 |
ENST00000388740.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr2_-_228028829 | 0.27 |
ENST00000396625.3
ENST00000329662.7 |
COL4A4
|
collagen, type IV, alpha 4 |
| chr2_+_236578248 | 0.27 |
ENST00000409538.1
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr11_+_7618413 | 0.27 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr19_-_10047219 | 0.27 |
ENST00000264833.4
|
OLFM2
|
olfactomedin 2 |
| chr22_-_46373004 | 0.27 |
ENST00000339464.4
|
WNT7B
|
wingless-type MMTV integration site family, member 7B |
| chr14_-_106805716 | 0.27 |
ENST00000438142.2
|
IGHV4-31
|
immunoglobulin heavy variable 4-31 |
| chr5_+_5140436 | 0.27 |
ENST00000511368.1
ENST00000274181.7 |
ADAMTS16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16 |
| chr6_-_47277634 | 0.27 |
ENST00000296861.2
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr16_-_8962853 | 0.26 |
ENST00000565287.1
ENST00000311052.5 |
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa |
| chr17_+_76142434 | 0.26 |
ENST00000340363.5
ENST00000586999.1 |
C17orf99
|
chromosome 17 open reading frame 99 |
| chr12_-_53074182 | 0.26 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr18_-_24445664 | 0.26 |
ENST00000578776.1
|
AQP4
|
aquaporin 4 |
| chr8_+_106330920 | 0.26 |
ENST00000407775.2
|
ZFPM2
|
zinc finger protein, FOG family member 2 |
| chr11_-_417308 | 0.26 |
ENST00000397632.3
ENST00000382520.2 |
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr20_-_5426332 | 0.26 |
ENST00000420529.1
|
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chr19_-_42806842 | 0.26 |
ENST00000596265.1
|
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr2_-_233792837 | 0.26 |
ENST00000373552.4
ENST00000409079.1 |
NGEF
|
neuronal guanine nucleotide exchange factor |
| chr10_-_49459800 | 0.26 |
ENST00000305531.3
|
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr10_-_116286563 | 0.26 |
ENST00000369253.2
|
ABLIM1
|
actin binding LIM protein 1 |
| chr3_-_75834722 | 0.26 |
ENST00000471541.2
|
ZNF717
|
zinc finger protein 717 |
| chr3_-_47517302 | 0.25 |
ENST00000441517.2
ENST00000545718.1 |
SCAP
|
SREBF chaperone |
| chr16_-_11922665 | 0.25 |
ENST00000573319.1
ENST00000577041.1 ENST00000574028.1 ENST00000571259.1 ENST00000573037.1 ENST00000571158.1 |
BCAR4
|
breast cancer anti-estrogen resistance 4 (non-protein coding) |
| chr8_-_81083341 | 0.25 |
ENST00000519303.2
|
TPD52
|
tumor protein D52 |
| chr11_+_64004888 | 0.25 |
ENST00000541681.1
|
VEGFB
|
vascular endothelial growth factor B |
| chr1_+_220863187 | 0.25 |
ENST00000294889.5
|
C1orf115
|
chromosome 1 open reading frame 115 |
| chr22_-_32651326 | 0.25 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr2_-_239140276 | 0.25 |
ENST00000334973.4
|
AC016757.3
|
Protein LOC151174 |
| chr16_+_2880254 | 0.25 |
ENST00000570670.1
|
ZG16B
|
zymogen granule protein 16B |
| chrX_-_102565932 | 0.25 |
ENST00000372674.1
ENST00000372677.3 |
BEX2
|
brain expressed X-linked 2 |
| chr10_-_118032697 | 0.25 |
ENST00000439649.3
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr17_-_39150385 | 0.25 |
ENST00000391586.1
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr22_-_30942669 | 0.25 |
ENST00000402034.2
|
SEC14L6
|
SEC14-like 6 (S. cerevisiae) |
| chr10_-_49732281 | 0.25 |
ENST00000374170.1
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr1_+_153651078 | 0.25 |
ENST00000368680.3
|
NPR1
|
natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A) |
| chr17_-_46507537 | 0.25 |
ENST00000336915.6
|
SKAP1
|
src kinase associated phosphoprotein 1 |
| chr2_+_74741569 | 0.25 |
ENST00000233638.7
|
TLX2
|
T-cell leukemia homeobox 2 |
| chr11_+_64107663 | 0.24 |
ENST00000356786.5
|
CCDC88B
|
coiled-coil domain containing 88B |
| chr15_-_42448788 | 0.24 |
ENST00000382396.4
ENST00000397272.3 |
PLA2G4F
|
phospholipase A2, group IVF |
| chr17_-_39203519 | 0.24 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr3_-_183543301 | 0.24 |
ENST00000318631.3
ENST00000431348.1 |
MAP6D1
|
MAP6 domain containing 1 |
| chr7_+_192969 | 0.24 |
ENST00000313766.5
|
FAM20C
|
family with sequence similarity 20, member C |
| chr4_+_41362796 | 0.24 |
ENST00000508501.1
ENST00000512946.1 ENST00000313860.7 ENST00000512632.1 ENST00000512820.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_+_33207381 | 0.24 |
ENST00000401073.2
|
KIAA1522
|
KIAA1522 |
| chr11_-_64570706 | 0.24 |
ENST00000294066.2
ENST00000377350.3 |
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr7_-_132261253 | 0.24 |
ENST00000321063.4
|
PLXNA4
|
plexin A4 |
| chr7_+_142982023 | 0.24 |
ENST00000359333.3
ENST00000409244.1 ENST00000409541.1 ENST00000410004.1 |
TMEM139
|
transmembrane protein 139 |
| chr3_-_190580404 | 0.24 |
ENST00000442080.1
|
GMNC
|
geminin coiled-coil domain containing |
| chr11_-_2170786 | 0.24 |
ENST00000300632.5
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr11_-_59612969 | 0.23 |
ENST00000541311.1
ENST00000257248.2 |
GIF
|
gastric intrinsic factor (vitamin B synthesis) |
| chr11_+_61447845 | 0.23 |
ENST00000257215.5
|
DAGLA
|
diacylglycerol lipase, alpha |
| chr19_-_50979981 | 0.23 |
ENST00000595790.1
ENST00000600100.1 |
FAM71E1
|
family with sequence similarity 71, member E1 |
| chr16_+_31539197 | 0.23 |
ENST00000564707.1
|
AHSP
|
alpha hemoglobin stabilizing protein |
| chr13_+_76334498 | 0.23 |
ENST00000534657.1
|
LMO7
|
LIM domain 7 |
| chr11_+_34643600 | 0.23 |
ENST00000530286.1
ENST00000533754.1 |
EHF
|
ets homologous factor |
| chr2_+_47596287 | 0.23 |
ENST00000263735.4
|
EPCAM
|
epithelial cell adhesion molecule |
Network of associatons between targets according to the STRING database.
First level regulatory network of ID4_TCF4_SNAI2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.8 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.4 | 1.1 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.3 | 0.8 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.2 | 0.6 | GO:0003064 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.2 | 0.6 | GO:0002194 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.2 | 0.5 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.2 | 0.5 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.2 | 0.5 | GO:0072019 | proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.2 | 0.6 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 0.6 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.1 | 0.7 | GO:0009439 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.1 | 0.7 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.6 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.5 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 0.4 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.1 | 0.4 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.1 | 0.4 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.1 | 0.7 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 1.2 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.3 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) regulation of chromatin silencing at telomere(GO:0031938) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.3 | GO:0051796 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.1 | 0.3 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.8 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.3 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.1 | 0.3 | GO:0050894 | determination of affect(GO:0050894) |
| 0.1 | 0.3 | GO:0071848 | circadian temperature homeostasis(GO:0060086) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.1 | 0.3 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.1 | 0.3 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 0.6 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 0.3 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.1 | 0.4 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.1 | 0.4 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.3 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.1 | 0.4 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 0.1 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.1 | 0.6 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.5 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.7 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.2 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.1 | 0.5 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.1 | 0.1 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.1 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 0.2 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.1 | 0.3 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 0.2 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.1 | 0.2 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.1 | 0.2 | GO:1905166 | negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.3 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 0.2 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.1 | 1.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 0.2 | GO:0032900 | negative regulation of neurotrophin production(GO:0032900) |
| 0.1 | 0.4 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.1 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 0.1 | GO:1902908 | regulation of melanosome transport(GO:1902908) |
| 0.1 | 0.2 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 0.1 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.1 | 0.5 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.3 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.6 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.4 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.2 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.1 | GO:0072554 | blood vessel lumenization(GO:0072554) |
| 0.1 | 0.2 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.1 | 0.2 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.1 | 0.4 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.1 | 0.2 | GO:1904481 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.1 | 0.2 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.1 | 0.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.4 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.3 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 0.3 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.5 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.2 | GO:0010840 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.1 | 0.4 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.1 | 0.2 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.1 | 0.2 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.1 | 0.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.4 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.2 | GO:0043000 | Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.1 | 0.5 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.2 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.6 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.2 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.1 | 0.2 | GO:0055099 | detection of endogenous stimulus(GO:0009726) response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.2 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 0.2 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.1 | 0.2 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.1 | 0.3 | GO:0072249 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.1 | 0.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.1 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.1 | 0.2 | GO:1990641 | response to iron ion starvation(GO:1990641) |
| 0.1 | 0.2 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.1 | 0.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.4 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.0 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.0 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.0 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.2 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.1 | GO:2000775 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) regulation of chromosome condensation(GO:0060623) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.0 | 0.2 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.0 | GO:0032899 | neurotrophin production(GO:0032898) regulation of neurotrophin production(GO:0032899) |
| 0.0 | 0.2 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.0 | 0.1 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.2 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.2 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.0 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.2 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.4 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.1 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.0 | 0.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.1 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.0 | 0.6 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.2 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 | 0.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.2 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.2 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.2 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.3 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.0 | 0.1 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.0 | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.0 | 0.1 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.3 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.1 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.0 | 0.1 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.0 | 0.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.0 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.0 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.1 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 | 0.2 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.0 | 0.1 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.2 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 1.1 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.3 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.1 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 | 0.1 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.2 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.1 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.0 | 0.1 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.2 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:1902623 | complement component C5a signaling pathway(GO:0038178) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.2 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.0 | GO:0061317 | canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.2 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.1 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.0 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.2 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.2 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.1 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.2 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.3 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.4 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.3 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.0 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.0 | 0.1 | GO:1904347 | positive regulation of eating behavior(GO:1904000) regulation of small intestine smooth muscle contraction(GO:1904347) positive regulation of small intestine smooth muscle contraction(GO:1904349) small intestine smooth muscle contraction(GO:1990770) |
| 0.0 | 0.1 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.0 | 0.2 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.3 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.1 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 0.1 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0010041 | response to iron(III) ion(GO:0010041) |
| 0.0 | 0.1 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.6 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.3 | GO:0048296 | regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.0 | 0.8 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.1 | GO:0001172 | transcription, RNA-templated(GO:0001172) negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.1 | GO:0002268 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.1 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.2 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) |
| 0.0 | 0.2 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.1 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.0 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.3 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.2 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.0 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.5 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0003266 | regulation of secondary heart field cardioblast proliferation(GO:0003266) |
| 0.0 | 0.2 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.5 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.0 | 0.1 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.0 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.5 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.0 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.1 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.2 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.0 | 0.6 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.1 | GO:1902617 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) response to fluoride(GO:1902617) |
| 0.0 | 0.1 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.0 | 0.1 | GO:0071899 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.0 | 0.1 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.0 | 0.1 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.1 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.1 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.2 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.6 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.2 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.2 | GO:1904729 | regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.1 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.0 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.0 | 0.0 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 2.0 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.0 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.0 | 0.4 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.5 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.2 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.8 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.0 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.1 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 1.7 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.1 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.2 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 0.3 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.1 | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.0 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.0 | 0.0 | GO:0034136 | negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) |
| 0.0 | 0.1 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.2 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.0 | GO:0048320 | axial mesoderm morphogenesis(GO:0048319) axial mesoderm formation(GO:0048320) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.0 | GO:0007497 | posterior midgut development(GO:0007497) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.1 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.1 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.0 | 0.1 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.0 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.0 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 0.2 | GO:0042748 | circadian sleep/wake cycle, non-REM sleep(GO:0042748) |
| 0.0 | 0.4 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.0 | 0.4 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.2 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.3 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.9 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.1 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.0 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.0 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.3 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.1 | GO:0035698 | CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.0 | 0.2 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.0 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.1 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.0 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.0 | GO:0072134 | nephrogenic mesenchyme morphogenesis(GO:0072134) |
| 0.0 | 0.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.0 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.1 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.0 | 0.1 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:1900040 | regulation of interleukin-2 secretion(GO:1900040) positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.2 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.1 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 | 0.1 | GO:0050823 | peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.0 | 0.0 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.0 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.0 | 2.9 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.0 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.0 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.0 | 0.0 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.2 | GO:0071415 | cellular response to purine-containing compound(GO:0071415) |
| 0.0 | 0.3 | GO:0030826 | regulation of cGMP biosynthetic process(GO:0030826) |
| 0.0 | 0.1 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.0 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.6 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.0 | 0.0 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.0 | 0.0 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.1 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 0.1 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.0 | 0.4 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.0 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0016128 | phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.4 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.0 | 0.0 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.0 | GO:2000669 | dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.0 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.0 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.0 | 0.0 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.0 | 0.1 | GO:0010813 | neuropeptide catabolic process(GO:0010813) substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.1 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.3 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.2 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.0 | 0.0 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.1 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 1.1 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.0 | 0.1 | GO:1904044 | response to aldosterone(GO:1904044) |
| 0.0 | 0.1 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.2 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.0 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.0 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.1 | GO:0042182 | ketone catabolic process(GO:0042182) |
| 0.0 | 0.0 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.0 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.0 | GO:1902567 | negative regulation of eosinophil activation(GO:1902567) |
| 0.0 | 0.1 | GO:0032445 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.1 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 | 0.6 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.3 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 0.0 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.1 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.2 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.0 | GO:0072708 | response to sorbitol(GO:0072708) cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0033239 | negative regulation of cellular amine metabolic process(GO:0033239) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.4 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.0 | GO:0042137 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.0 | GO:0006550 | isoleucine metabolic process(GO:0006549) isoleucine catabolic process(GO:0006550) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.0 | 0.0 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.0 | 0.2 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.3 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.0 | GO:0045953 | negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.0 | 0.0 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.1 | GO:1904995 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.0 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.0 | 0.2 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.1 | GO:0009111 | vitamin catabolic process(GO:0009111) |
| 0.0 | 0.1 | GO:0010635 | regulation of mitochondrial fusion(GO:0010635) |
| 0.0 | 0.1 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) |
| 0.0 | 0.6 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.0 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.0 | GO:0090155 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.0 | GO:1902995 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.0 | 0.0 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 0.0 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.1 | GO:0044146 | negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.0 | 0.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.0 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.1 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.0 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 0.0 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.0 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.0 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.0 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.0 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.0 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.0 | GO:0033967 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.0 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) positive regulation of monocyte extravasation(GO:2000439) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.1 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.0 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 0.1 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.2 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.0 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.1 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.3 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.3 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.0 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 0.8 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.4 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.4 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 0.3 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 1.0 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 1.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.8 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.3 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.1 | 0.4 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 1.0 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 0.5 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 0.4 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 0.2 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.1 | 0.6 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.2 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.0 | 0.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.0 | 4.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.4 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.2 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.2 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0031261 | GINS complex(GO:0000811) DNA replication preinitiation complex(GO:0031261) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 1.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0032173 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.0 | 0.1 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.0 | 0.0 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 2.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 3.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.0 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.0 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.0 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.0 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.3 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.5 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.0 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.0 | 0.1 | GO:0036338 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.0 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.0 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 0.6 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.2 | 0.8 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.4 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.1 | 0.6 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.1 | 0.6 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.1 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.4 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 0.6 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 0.4 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 0.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.4 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.1 | 1.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.3 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.1 | 0.4 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 0.3 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.3 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.1 | 0.6 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 0.2 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.6 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.3 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.4 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.1 | 0.8 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.2 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.1 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.2 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.3 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.3 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.4 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 2.1 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.0 | 0.2 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0016775 | phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.4 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0000994 | RNA polymerase III core binding(GO:0000994) |
| 0.0 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0051990 | (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.0 | 0.0 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 1.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.1 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.2 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.0 | 0.1 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.0 | 0.4 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.0 | 0.1 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.2 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.1 | GO:0038047 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 1.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.3 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.1 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 0.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.2 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.1 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.2 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.5 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.0 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.1 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.7 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.0 | 0.3 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.5 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 1.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.1 | GO:0047726 | iron-cytochrome-c reductase activity(GO:0047726) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.0 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.0 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.0 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.1 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.0 | 0.0 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.0 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.0 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.3 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.0 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.0 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.5 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.0 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.0 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.0 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.0 | 0.0 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.1 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.0 | 0.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.0 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.0 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.0 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.0 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.0 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.0 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.0 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.0 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.0 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.0 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.0 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.2 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 0.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 0.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.0 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.2 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.4 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.0 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 6.9 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 1.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 0.3 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.1 | 1.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.5 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.1 | 2.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 2.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.0 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.5 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.7 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.0 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.1 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.0 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.0 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.6 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |