Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
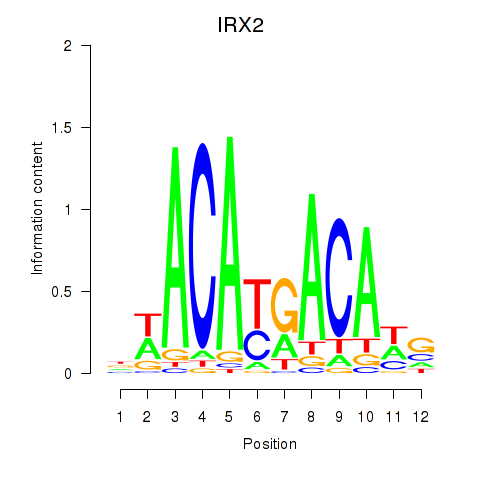
Results for IRX2
Z-value: 0.46
Transcription factors associated with IRX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX2
|
ENSG00000170561.8 | iroquois homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX2 | hg19_v2_chr5_-_2751762_2751784 | 0.30 | 1.5e-01 | Click! |
Activity profile of IRX2 motif
Sorted Z-values of IRX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_80263506 | 0.61 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr5_+_147258266 | 0.60 |
ENST00000296694.4
|
SCGB3A2
|
secretoglobin, family 3A, member 2 |
| chr3_-_127441406 | 0.55 |
ENST00000487473.1
ENST00000484451.1 |
MGLL
|
monoglyceride lipase |
| chrX_+_9431324 | 0.41 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr1_+_10292308 | 0.37 |
ENST00000377081.1
|
KIF1B
|
kinesin family member 1B |
| chr11_+_103907308 | 0.33 |
ENST00000302259.3
|
DDI1
|
DNA-damage inducible 1 homolog 1 (S. cerevisiae) |
| chr10_-_61122220 | 0.32 |
ENST00000422313.2
ENST00000435852.2 ENST00000442566.3 ENST00000373868.2 ENST00000277705.6 ENST00000373867.3 ENST00000419214.2 |
FAM13C
|
family with sequence similarity 13, member C |
| chr4_-_153332886 | 0.31 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr6_-_53013620 | 0.31 |
ENST00000259803.7
|
GCM1
|
glial cells missing homolog 1 (Drosophila) |
| chr5_+_54320078 | 0.31 |
ENST00000231009.2
|
GZMK
|
granzyme K (granzyme 3; tryptase II) |
| chr1_+_74701062 | 0.25 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chrX_-_49042778 | 0.24 |
ENST00000538114.1
ENST00000376310.3 ENST00000376317.3 ENST00000417014.1 |
PRICKLE3
|
prickle homolog 3 (Drosophila) |
| chr17_-_2614927 | 0.24 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chrX_-_21676442 | 0.23 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr5_-_35048047 | 0.23 |
ENST00000231420.6
|
AGXT2
|
alanine--glyoxylate aminotransferase 2 |
| chr4_+_57371509 | 0.22 |
ENST00000360096.2
|
ARL9
|
ADP-ribosylation factor-like 9 |
| chr11_+_33037652 | 0.22 |
ENST00000311388.3
|
DEPDC7
|
DEP domain containing 7 |
| chr3_+_97887544 | 0.22 |
ENST00000356526.2
|
OR5H15
|
olfactory receptor, family 5, subfamily H, member 15 |
| chr14_+_29236269 | 0.20 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr1_-_207095324 | 0.20 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr8_+_40018977 | 0.19 |
ENST00000520487.1
|
RP11-470M17.2
|
RP11-470M17.2 |
| chr19_-_35719609 | 0.19 |
ENST00000324675.3
|
FAM187B
|
family with sequence similarity 187, member B |
| chr12_-_16758304 | 0.19 |
ENST00000320122.6
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr1_-_157811588 | 0.19 |
ENST00000368174.4
|
CD5L
|
CD5 molecule-like |
| chr4_-_168155417 | 0.19 |
ENST00000511269.1
ENST00000506697.1 ENST00000512042.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chrX_+_30261847 | 0.19 |
ENST00000378981.3
ENST00000397550.1 |
MAGEB1
|
melanoma antigen family B, 1 |
| chr12_-_16758059 | 0.18 |
ENST00000261169.6
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr11_+_62648336 | 0.18 |
ENST00000338663.7
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr19_+_16308711 | 0.18 |
ENST00000429941.2
ENST00000444449.2 ENST00000589822.1 |
AP1M1
|
adaptor-related protein complex 1, mu 1 subunit |
| chr6_-_31620455 | 0.17 |
ENST00000437771.1
ENST00000404765.2 ENST00000375964.6 ENST00000211379.5 |
BAG6
|
BCL2-associated athanogene 6 |
| chr21_-_31538971 | 0.17 |
ENST00000286808.3
|
CLDN17
|
claudin 17 |
| chr18_-_33702078 | 0.17 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr5_-_94417314 | 0.17 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr4_-_70505358 | 0.17 |
ENST00000457664.2
ENST00000604629.1 ENST00000604021.1 |
UGT2A2
|
UDP glucuronosyltransferase 2 family, polypeptide A2 |
| chr16_+_21244986 | 0.16 |
ENST00000311620.5
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr3_-_151102529 | 0.16 |
ENST00000302632.3
|
P2RY12
|
purinergic receptor P2Y, G-protein coupled, 12 |
| chr12_+_32655110 | 0.16 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr12_+_66218903 | 0.16 |
ENST00000393577.3
|
HMGA2
|
high mobility group AT-hook 2 |
| chr6_+_88117683 | 0.15 |
ENST00000369562.4
|
C6ORF165
|
UPF0704 protein C6orf165 |
| chr5_-_94417339 | 0.15 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr6_-_31620403 | 0.15 |
ENST00000451898.1
ENST00000439687.2 ENST00000362049.6 ENST00000424480.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr2_+_159651821 | 0.15 |
ENST00000309950.3
ENST00000409042.1 |
DAPL1
|
death associated protein-like 1 |
| chr4_-_80247162 | 0.15 |
ENST00000286794.4
|
NAA11
|
N(alpha)-acetyltransferase 11, NatA catalytic subunit |
| chr6_+_37897735 | 0.15 |
ENST00000373389.5
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr11_+_111126707 | 0.15 |
ENST00000280325.4
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chr1_-_238649319 | 0.15 |
ENST00000400946.2
|
RP11-371I1.2
|
long intergenic non-protein coding RNA 1139 |
| chr6_-_9933500 | 0.14 |
ENST00000492169.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr1_+_31883048 | 0.14 |
ENST00000536859.1
|
SERINC2
|
serine incorporator 2 |
| chrX_+_150869023 | 0.14 |
ENST00000448324.1
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr10_+_46997926 | 0.14 |
ENST00000374314.4
|
GPRIN2
|
G protein regulated inducer of neurite outgrowth 2 |
| chr21_+_43823983 | 0.13 |
ENST00000291535.6
ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr4_-_14889791 | 0.13 |
ENST00000509654.1
ENST00000515031.1 ENST00000505089.2 |
LINC00504
|
long intergenic non-protein coding RNA 504 |
| chr1_+_113009163 | 0.13 |
ENST00000256640.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr1_-_207095212 | 0.13 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr3_-_11645925 | 0.12 |
ENST00000413604.1
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr8_+_24151553 | 0.12 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr19_+_3762645 | 0.12 |
ENST00000330133.4
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr1_+_113010056 | 0.12 |
ENST00000369686.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr4_+_80748640 | 0.12 |
ENST00000514836.1
|
PCAT4
|
prostate cancer associated transcript 4 (non-protein coding) |
| chr18_+_5748793 | 0.12 |
ENST00000566533.1
ENST00000562452.2 |
RP11-945C19.1
|
RP11-945C19.1 |
| chr1_-_111506562 | 0.12 |
ENST00000485275.2
ENST00000369763.4 |
LRIF1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr10_-_100027943 | 0.12 |
ENST00000260702.3
|
LOXL4
|
lysyl oxidase-like 4 |
| chr14_+_75536280 | 0.11 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr16_+_68678892 | 0.11 |
ENST00000429102.2
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental) |
| chr3_-_112565703 | 0.11 |
ENST00000488794.1
|
CD200R1L
|
CD200 receptor 1-like |
| chr6_-_159466136 | 0.11 |
ENST00000367066.3
ENST00000326965.6 |
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr8_+_30244580 | 0.11 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chrX_-_100129128 | 0.11 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr11_-_59383617 | 0.10 |
ENST00000263847.1
|
OSBP
|
oxysterol binding protein |
| chrX_+_19373700 | 0.10 |
ENST00000379804.1
|
PDHA1
|
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr1_-_173793458 | 0.10 |
ENST00000356198.2
|
CENPL
|
centromere protein L |
| chr14_+_50999744 | 0.10 |
ENST00000441560.2
|
ATL1
|
atlastin GTPase 1 |
| chr15_-_20193370 | 0.10 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr7_-_107770794 | 0.10 |
ENST00000205386.4
ENST00000418464.1 ENST00000388781.3 ENST00000388780.3 ENST00000414450.2 |
LAMB4
|
laminin, beta 4 |
| chr14_+_75536335 | 0.10 |
ENST00000554763.1
ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr19_-_38720354 | 0.10 |
ENST00000416611.1
|
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr12_+_66218598 | 0.10 |
ENST00000541363.1
|
HMGA2
|
high mobility group AT-hook 2 |
| chr12_+_40787194 | 0.09 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr5_-_180242534 | 0.09 |
ENST00000333055.3
ENST00000513431.1 |
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr6_-_27841289 | 0.09 |
ENST00000355981.2
|
HIST1H4L
|
histone cluster 1, H4l |
| chr11_-_118305921 | 0.09 |
ENST00000532619.1
|
RP11-770J1.4
|
RP11-770J1.4 |
| chr12_+_54402790 | 0.09 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr5_+_167913450 | 0.09 |
ENST00000231572.3
ENST00000538719.1 |
RARS
|
arginyl-tRNA synthetase |
| chr3_-_122102065 | 0.09 |
ENST00000479899.1
ENST00000291458.5 ENST00000497726.1 |
CCDC58
|
coiled-coil domain containing 58 |
| chrX_+_155227371 | 0.09 |
ENST00000369423.2
ENST00000540897.1 |
IL9R
|
interleukin 9 receptor |
| chr1_-_156698181 | 0.09 |
ENST00000313146.6
|
ISG20L2
|
interferon stimulated exonuclease gene 20kDa-like 2 |
| chrX_+_155227246 | 0.08 |
ENST00000244174.5
ENST00000424344.3 |
IL9R
|
interleukin 9 receptor |
| chr16_-_4401258 | 0.08 |
ENST00000577031.1
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr14_-_106312010 | 0.08 |
ENST00000390556.2
|
IGHD
|
immunoglobulin heavy constant delta |
| chr5_-_142814241 | 0.08 |
ENST00000504572.1
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr6_+_27925019 | 0.08 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr3_-_167371740 | 0.08 |
ENST00000466760.1
ENST00000479765.1 |
WDR49
|
WD repeat domain 49 |
| chr3_-_186262166 | 0.08 |
ENST00000307944.5
|
CRYGS
|
crystallin, gamma S |
| chr20_+_56136136 | 0.08 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr1_+_29138654 | 0.08 |
ENST00000234961.2
|
OPRD1
|
opioid receptor, delta 1 |
| chr17_+_900342 | 0.08 |
ENST00000327158.4
|
TIMM22
|
translocase of inner mitochondrial membrane 22 homolog (yeast) |
| chr6_-_159466042 | 0.07 |
ENST00000338313.5
|
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr4_+_189060573 | 0.07 |
ENST00000332517.3
|
TRIML1
|
tripartite motif family-like 1 |
| chr8_-_82359662 | 0.07 |
ENST00000519260.1
ENST00000256103.2 |
PMP2
|
peripheral myelin protein 2 |
| chr1_-_190446759 | 0.07 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr11_-_76381029 | 0.07 |
ENST00000407242.2
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr1_+_215747118 | 0.07 |
ENST00000448333.1
|
KCTD3
|
potassium channel tetramerization domain containing 3 |
| chr19_-_40336969 | 0.06 |
ENST00000599134.1
ENST00000597634.1 ENST00000598417.1 ENST00000601274.1 ENST00000594309.1 ENST00000221801.3 |
FBL
|
fibrillarin |
| chr12_+_4714145 | 0.06 |
ENST00000545342.1
|
DYRK4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chrX_-_24045303 | 0.06 |
ENST00000328046.8
|
KLHL15
|
kelch-like family member 15 |
| chr19_+_18669809 | 0.06 |
ENST00000602094.1
|
KXD1
|
KxDL motif containing 1 |
| chr10_+_102672712 | 0.06 |
ENST00000370271.3
ENST00000370269.3 ENST00000609386.1 |
FAM178A
|
family with sequence similarity 178, member A |
| chr2_+_242577097 | 0.06 |
ENST00000419606.1
ENST00000474739.2 ENST00000396411.3 ENST00000425239.1 ENST00000400771.3 ENST00000430617.2 |
ATG4B
|
autophagy related 4B, cysteine peptidase |
| chr7_+_95115210 | 0.06 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr3_+_108541545 | 0.06 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr12_+_128399965 | 0.06 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr18_+_55018044 | 0.06 |
ENST00000324000.3
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr3_+_132843652 | 0.06 |
ENST00000508711.1
|
TMEM108
|
transmembrane protein 108 |
| chr1_+_166958346 | 0.06 |
ENST00000367872.4
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr2_+_168675182 | 0.05 |
ENST00000305861.1
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chrX_-_100129320 | 0.05 |
ENST00000372966.3
|
NOX1
|
NADPH oxidase 1 |
| chr17_-_3337135 | 0.05 |
ENST00000248384.1
|
OR1E2
|
olfactory receptor, family 1, subfamily E, member 2 |
| chr22_-_36013368 | 0.05 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr7_-_14880892 | 0.05 |
ENST00000406247.3
ENST00000399322.3 ENST00000258767.5 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr6_+_24775153 | 0.05 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr2_-_109605663 | 0.05 |
ENST00000409271.1
ENST00000258443.2 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr3_+_46616017 | 0.05 |
ENST00000542931.1
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr1_+_166958497 | 0.05 |
ENST00000367870.2
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr10_-_28270795 | 0.05 |
ENST00000545014.1
|
ARMC4
|
armadillo repeat containing 4 |
| chr12_-_91505608 | 0.05 |
ENST00000266718.4
|
LUM
|
lumican |
| chr19_-_29704448 | 0.05 |
ENST00000304863.4
|
UQCRFS1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr16_-_5083917 | 0.04 |
ENST00000312251.3
ENST00000381955.3 |
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr2_+_196313239 | 0.04 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr10_+_134150835 | 0.04 |
ENST00000432555.2
|
LRRC27
|
leucine rich repeat containing 27 |
| chr2_-_98280383 | 0.04 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr4_+_88896819 | 0.04 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr1_-_151148442 | 0.04 |
ENST00000441701.1
ENST00000416280.2 |
TMOD4
|
tropomodulin 4 (muscle) |
| chr10_+_1095416 | 0.04 |
ENST00000358220.1
|
WDR37
|
WD repeat domain 37 |
| chr14_-_65289812 | 0.04 |
ENST00000389720.3
ENST00000389721.5 ENST00000389722.3 |
SPTB
|
spectrin, beta, erythrocytic |
| chr2_-_233877912 | 0.04 |
ENST00000264051.3
|
NGEF
|
neuronal guanine nucleotide exchange factor |
| chr10_-_4285923 | 0.04 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr19_-_38720294 | 0.04 |
ENST00000412732.1
ENST00000456296.1 |
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr8_+_94767072 | 0.04 |
ENST00000452276.1
ENST00000453321.3 ENST00000498673.1 ENST00000518319.1 |
TMEM67
|
transmembrane protein 67 |
| chr6_-_136847099 | 0.03 |
ENST00000438100.2
|
MAP7
|
microtubule-associated protein 7 |
| chr4_-_109541539 | 0.03 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr1_-_173572181 | 0.03 |
ENST00000536496.1
ENST00000367714.3 |
SLC9C2
|
solute carrier family 9, member C2 (putative) |
| chr6_+_46661575 | 0.03 |
ENST00000450697.1
|
TDRD6
|
tudor domain containing 6 |
| chr3_-_194072019 | 0.03 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chr17_-_7761256 | 0.03 |
ENST00000575208.1
|
LSMD1
|
LSM domain containing 1 |
| chrX_-_132095419 | 0.03 |
ENST00000370836.2
ENST00000521489.1 |
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr6_-_136847610 | 0.03 |
ENST00000454590.1
ENST00000432797.2 |
MAP7
|
microtubule-associated protein 7 |
| chr18_+_50278430 | 0.03 |
ENST00000578080.1
ENST00000582875.1 ENST00000412726.1 |
DCC
|
deleted in colorectal carcinoma |
| chr20_+_43849941 | 0.03 |
ENST00000372769.3
|
SEMG2
|
semenogelin II |
| chr1_+_67673297 | 0.03 |
ENST00000425614.1
ENST00000395227.1 |
IL23R
|
interleukin 23 receptor |
| chr8_+_67405794 | 0.03 |
ENST00000522977.1
ENST00000480005.1 |
C8orf46
|
chromosome 8 open reading frame 46 |
| chr4_+_114214125 | 0.03 |
ENST00000509550.1
|
ANK2
|
ankyrin 2, neuronal |
| chr16_+_69373323 | 0.03 |
ENST00000254940.5
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr17_-_26127525 | 0.02 |
ENST00000313735.6
|
NOS2
|
nitric oxide synthase 2, inducible |
| chr4_+_158141843 | 0.02 |
ENST00000509417.1
ENST00000296526.7 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr7_-_33140498 | 0.02 |
ENST00000448915.1
|
RP9
|
retinitis pigmentosa 9 (autosomal dominant) |
| chr4_+_158141899 | 0.02 |
ENST00000264426.9
ENST00000506284.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr11_+_59824127 | 0.02 |
ENST00000278865.3
|
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr1_+_110527308 | 0.02 |
ENST00000369799.5
|
AHCYL1
|
adenosylhomocysteinase-like 1 |
| chr7_+_150065278 | 0.02 |
ENST00000519397.1
ENST00000479668.1 ENST00000540729.1 |
REPIN1
|
replication initiator 1 |
| chr19_-_42463418 | 0.02 |
ENST00000600292.1
ENST00000601078.1 ENST00000601891.1 ENST00000222008.6 |
RABAC1
|
Rab acceptor 1 (prenylated) |
| chr16_+_58426296 | 0.02 |
ENST00000426538.2
ENST00000328514.7 ENST00000318129.5 |
GINS3
|
GINS complex subunit 3 (Psf3 homolog) |
| chr2_+_27851863 | 0.02 |
ENST00000264718.3
ENST00000610189.1 |
GPN1
|
GPN-loop GTPase 1 |
| chr14_+_52327109 | 0.02 |
ENST00000335281.4
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr1_-_151148492 | 0.02 |
ENST00000295314.4
|
TMOD4
|
tropomodulin 4 (muscle) |
| chr6_+_30908747 | 0.02 |
ENST00000462446.1
ENST00000304311.2 |
DPCR1
|
diffuse panbronchiolitis critical region 1 |
| chr4_+_159131630 | 0.02 |
ENST00000504569.1
ENST00000509278.1 ENST00000514558.1 ENST00000503200.1 |
TMEM144
|
transmembrane protein 144 |
| chr12_+_66218212 | 0.02 |
ENST00000393578.3
ENST00000425208.2 ENST00000536545.1 ENST00000354636.3 |
HMGA2
|
high mobility group AT-hook 2 |
| chr19_+_13134772 | 0.01 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr16_+_81272287 | 0.01 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr7_-_105332084 | 0.01 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr2_-_70520539 | 0.01 |
ENST00000482975.2
ENST00000438261.1 |
SNRPG
|
small nuclear ribonucleoprotein polypeptide G |
| chrX_-_70331298 | 0.01 |
ENST00000456850.2
ENST00000473378.1 ENST00000487883.1 ENST00000374202.2 |
IL2RG
|
interleukin 2 receptor, gamma |
| chr5_-_138210977 | 0.01 |
ENST00000274711.6
ENST00000521094.2 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr7_+_80275953 | 0.01 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr2_-_166060571 | 0.01 |
ENST00000360093.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr14_+_23790655 | 0.01 |
ENST00000397276.2
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr1_+_152691998 | 0.01 |
ENST00000368775.2
|
C1orf68
|
chromosome 1 open reading frame 68 |
| chr2_-_89160770 | 0.01 |
ENST00000390240.2
|
IGKJ3
|
immunoglobulin kappa joining 3 |
| chr11_+_98891797 | 0.01 |
ENST00000527185.1
ENST00000528682.1 ENST00000524871.1 |
CNTN5
|
contactin 5 |
| chr3_+_148457585 | 0.01 |
ENST00000402260.1
|
AGTR1
|
angiotensin II receptor, type 1 |
| chr1_+_204839959 | 0.01 |
ENST00000404076.1
|
NFASC
|
neurofascin |
| chr4_+_187187337 | 0.01 |
ENST00000492972.2
|
F11
|
coagulation factor XI |
| chr18_+_32455201 | 0.00 |
ENST00000590831.2
|
DTNA
|
dystrobrevin, alpha |
| chr12_+_128399917 | 0.00 |
ENST00000544645.1
|
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chrX_-_138790348 | 0.00 |
ENST00000414978.1
ENST00000519895.1 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr2_+_171034646 | 0.00 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr1_+_166958504 | 0.00 |
ENST00000447624.1
|
MAEL
|
maelstrom spermatogenic transposon silencer |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.1 | 0.3 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 0.3 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.1 | 0.5 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.2 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.1 | 0.3 | GO:0031049 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.1 | 0.3 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.4 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.2 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.0 | 0.3 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 0.2 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.1 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.1 | GO:0032773 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.0 | 0.2 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.1 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.2 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.1 | GO:1990258 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.6 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.0 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.2 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0030849 | autosome(GO:0030849) |
| 0.0 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.6 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.0 | 0.2 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.1 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |