Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for IRX3
Z-value: 0.78
Transcription factors associated with IRX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX3
|
ENSG00000177508.11 | iroquois homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX3 | hg19_v2_chr16_-_54320675_54320715 | 0.24 | 2.4e-01 | Click! |
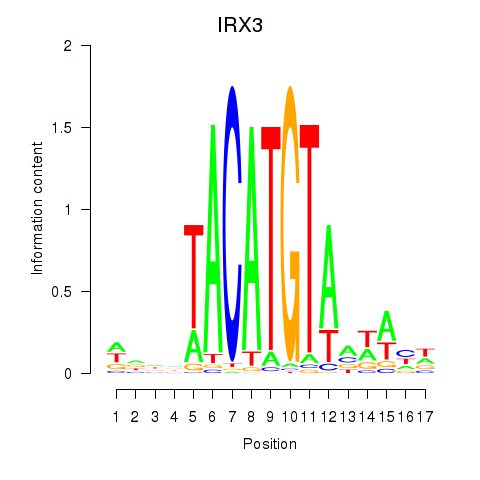
Activity profile of IRX3 motif
Sorted Z-values of IRX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_127541194 | 1.96 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase |
| chr5_+_54320078 | 1.66 |
ENST00000231009.2
|
GZMK
|
granzyme K (granzyme 3; tryptase II) |
| chr12_-_71182695 | 1.42 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr12_+_117348742 | 1.40 |
ENST00000309909.5
ENST00000455858.2 |
FBXW8
|
F-box and WD repeat domain containing 8 |
| chr10_+_118305435 | 1.36 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr5_+_35856951 | 1.32 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr4_-_70080449 | 1.24 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr4_-_76957214 | 1.22 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr1_+_87012753 | 1.21 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr3_+_122399444 | 1.07 |
ENST00000474629.2
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr11_+_35201826 | 1.04 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr7_-_92777606 | 1.03 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr3_-_127441406 | 1.02 |
ENST00000487473.1
ENST00000484451.1 |
MGLL
|
monoglyceride lipase |
| chr2_-_89545079 | 1.01 |
ENST00000468494.1
|
IGKV2-30
|
immunoglobulin kappa variable 2-30 |
| chr1_-_89488510 | 1.00 |
ENST00000564665.1
ENST00000370481.4 |
GBP3
|
guanylate binding protein 3 |
| chr3_-_123512688 | 0.98 |
ENST00000475616.1
|
MYLK
|
myosin light chain kinase |
| chr11_+_35211429 | 0.94 |
ENST00000525688.1
ENST00000278385.6 ENST00000533222.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr1_-_201438282 | 0.86 |
ENST00000367311.3
ENST00000367309.1 |
PHLDA3
|
pleckstrin homology-like domain, family A, member 3 |
| chr4_+_74606223 | 0.82 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr17_-_34207295 | 0.82 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr13_+_97928395 | 0.82 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr22_-_36556821 | 0.80 |
ENST00000531095.1
ENST00000397293.2 ENST00000349314.2 |
APOL3
|
apolipoprotein L, 3 |
| chr10_-_90712520 | 0.80 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr15_+_67420441 | 0.78 |
ENST00000558894.1
|
SMAD3
|
SMAD family member 3 |
| chr21_-_43528548 | 0.76 |
ENST00000329015.2
|
C21orf128
|
chromosome 21 open reading frame 128 |
| chr10_+_91092241 | 0.75 |
ENST00000371811.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr2_+_89975669 | 0.74 |
ENST00000474213.1
|
IGKV2D-30
|
immunoglobulin kappa variable 2D-30 |
| chr11_+_20044600 | 0.73 |
ENST00000311043.8
|
NAV2
|
neuron navigator 2 |
| chr15_+_72947079 | 0.70 |
ENST00000421285.3
|
GOLGA6B
|
golgin A6 family, member B |
| chr5_-_59481406 | 0.65 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr5_+_135385202 | 0.65 |
ENST00000514554.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr2_-_216878305 | 0.62 |
ENST00000263268.6
|
MREG
|
melanoregulin |
| chr3_-_122283079 | 0.61 |
ENST00000471785.1
ENST00000466126.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr13_-_88323218 | 0.61 |
ENST00000436290.2
ENST00000453832.2 ENST00000606590.1 |
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr17_+_25958174 | 0.59 |
ENST00000313648.6
ENST00000577392.1 ENST00000584661.1 ENST00000413914.2 |
LGALS9
|
lectin, galactoside-binding, soluble, 9 |
| chr17_+_18380051 | 0.58 |
ENST00000581545.1
ENST00000582333.1 ENST00000328114.6 ENST00000412421.2 ENST00000583322.1 ENST00000584941.1 |
LGALS9C
|
lectin, galactoside-binding, soluble, 9C |
| chr3_+_122283064 | 0.58 |
ENST00000296161.4
|
DTX3L
|
deltex 3-like (Drosophila) |
| chr8_-_125577940 | 0.56 |
ENST00000519168.1
ENST00000395508.2 |
MTSS1
|
metastasis suppressor 1 |
| chr5_+_175490540 | 0.56 |
ENST00000515817.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr2_-_207629997 | 0.55 |
ENST00000454776.2
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr5_+_147443534 | 0.55 |
ENST00000398454.1
ENST00000359874.3 ENST00000508733.1 ENST00000256084.7 |
SPINK5
|
serine peptidase inhibitor, Kazal type 5 |
| chr5_-_13944652 | 0.54 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr19_+_20959098 | 0.54 |
ENST00000360204.5
ENST00000594534.1 |
ZNF66
|
zinc finger protein 66 |
| chr7_-_41742697 | 0.53 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr2_-_16804320 | 0.53 |
ENST00000355549.2
|
FAM49A
|
family with sequence similarity 49, member A |
| chr4_-_95264008 | 0.51 |
ENST00000295256.5
|
HPGDS
|
hematopoietic prostaglandin D synthase |
| chr7_+_55086703 | 0.51 |
ENST00000455089.1
ENST00000342916.3 ENST00000344576.2 ENST00000420316.2 |
EGFR
|
epidermal growth factor receptor |
| chr7_-_122339162 | 0.50 |
ENST00000340112.2
|
RNF133
|
ring finger protein 133 |
| chr1_-_197115818 | 0.50 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr2_-_231989808 | 0.49 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr5_+_159848807 | 0.49 |
ENST00000352433.5
|
PTTG1
|
pituitary tumor-transforming 1 |
| chr1_+_87458692 | 0.49 |
ENST00000370548.2
ENST00000356813.4 |
RP5-1052I5.2
HS2ST1
|
Heparan sulfate 2-O-sulfotransferase 1 heparan sulfate 2-O-sulfotransferase 1 |
| chr5_-_177207634 | 0.48 |
ENST00000513554.1
ENST00000440605.3 |
FAM153A
|
family with sequence similarity 153, member A |
| chr6_-_35765088 | 0.48 |
ENST00000259938.2
|
CLPS
|
colipase, pancreatic |
| chr4_+_122722466 | 0.48 |
ENST00000243498.5
ENST00000379663.3 ENST00000509800.1 |
EXOSC9
|
exosome component 9 |
| chr1_+_196857144 | 0.47 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr2_-_207630033 | 0.47 |
ENST00000449792.1
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr4_-_104119528 | 0.46 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr5_+_110427983 | 0.46 |
ENST00000513710.2
ENST00000505303.1 |
WDR36
|
WD repeat domain 36 |
| chr2_-_231090344 | 0.46 |
ENST00000540870.1
ENST00000416610.1 |
SP110
|
SP110 nuclear body protein |
| chr15_+_75575176 | 0.45 |
ENST00000434739.3
|
GOLGA6D
|
golgin A6 family, member D |
| chr3_+_173116225 | 0.45 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chrX_-_15353629 | 0.45 |
ENST00000333590.4
ENST00000428964.1 ENST00000542278.1 |
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A |
| chr8_+_95558771 | 0.44 |
ENST00000391679.1
|
AC023632.1
|
HCG2009141; PRO2397; Uncharacterized protein |
| chr7_+_20687017 | 0.44 |
ENST00000258738.6
|
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr18_+_45778672 | 0.44 |
ENST00000600091.1
|
AC091150.1
|
HCG1818186; Uncharacterized protein |
| chr18_+_9885961 | 0.44 |
ENST00000306084.6
|
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa) |
| chr1_+_31883048 | 0.44 |
ENST00000536859.1
|
SERINC2
|
serine incorporator 2 |
| chr17_+_56232494 | 0.44 |
ENST00000268912.5
|
OR4D1
|
olfactory receptor, family 4, subfamily D, member 1 |
| chr22_+_30821732 | 0.43 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr7_-_101212244 | 0.43 |
ENST00000451953.1
ENST00000434537.1 ENST00000437900.1 |
LINC01007
|
long intergenic non-protein coding RNA 1007 |
| chr14_+_73563735 | 0.43 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25 |
| chr3_+_171561127 | 0.42 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr4_-_70826725 | 0.42 |
ENST00000353151.3
|
CSN2
|
casein beta |
| chrX_+_15767971 | 0.42 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr11_-_104905840 | 0.42 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr3_-_27498235 | 0.41 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr8_+_12803176 | 0.40 |
ENST00000524591.2
|
KIAA1456
|
KIAA1456 |
| chr12_-_91574142 | 0.40 |
ENST00000547937.1
|
DCN
|
decorin |
| chr5_+_159848854 | 0.39 |
ENST00000517480.1
ENST00000520452.1 ENST00000393964.1 |
PTTG1
|
pituitary tumor-transforming 1 |
| chr22_+_37309662 | 0.38 |
ENST00000403662.3
ENST00000262825.5 |
CSF2RB
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr3_+_173302222 | 0.38 |
ENST00000361589.4
|
NLGN1
|
neuroligin 1 |
| chr11_-_57177586 | 0.38 |
ENST00000529411.1
|
RP11-872D17.8
|
Uncharacterized protein |
| chr11_+_111783450 | 0.38 |
ENST00000537382.1
|
HSPB2
|
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr4_+_175839506 | 0.37 |
ENST00000505141.1
ENST00000359240.3 ENST00000445694.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr16_+_19421803 | 0.37 |
ENST00000541464.1
|
TMC5
|
transmembrane channel-like 5 |
| chr1_+_158985457 | 0.37 |
ENST00000567661.1
ENST00000474473.1 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr21_-_34186006 | 0.36 |
ENST00000490358.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr3_-_122283100 | 0.36 |
ENST00000492382.1
ENST00000462315.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr12_-_91573132 | 0.36 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr8_+_9009202 | 0.36 |
ENST00000518496.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chr12_-_87232644 | 0.36 |
ENST00000549405.2
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr19_-_10491234 | 0.35 |
ENST00000524462.1
ENST00000531836.1 ENST00000525621.1 |
TYK2
|
tyrosine kinase 2 |
| chr15_-_89089860 | 0.35 |
ENST00000558413.1
ENST00000564406.1 ENST00000268148.8 |
DET1
|
de-etiolated homolog 1 (Arabidopsis) |
| chr3_-_50378235 | 0.35 |
ENST00000357043.2
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1 |
| chr11_-_102496063 | 0.34 |
ENST00000260228.2
|
MMP20
|
matrix metallopeptidase 20 |
| chr7_+_123241908 | 0.34 |
ENST00000434204.1
ENST00000437535.1 ENST00000451215.1 |
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr2_+_233527443 | 0.34 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr2_+_133874577 | 0.33 |
ENST00000596384.1
|
AC011755.1
|
HCG2006742; Protein LOC100996685 |
| chr12_-_10251603 | 0.33 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr10_+_118350468 | 0.33 |
ENST00000358834.4
ENST00000528052.1 ENST00000442761.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr8_+_9009296 | 0.33 |
ENST00000521718.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chr3_-_191000172 | 0.33 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr5_+_68485433 | 0.32 |
ENST00000502689.1
|
CENPH
|
centromere protein H |
| chr17_-_29641084 | 0.32 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr10_+_118350522 | 0.32 |
ENST00000530319.1
ENST00000527980.1 ENST00000471549.1 ENST00000534537.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr3_+_142838091 | 0.32 |
ENST00000309575.3
|
CHST2
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 |
| chr19_+_38779778 | 0.32 |
ENST00000590738.1
ENST00000587519.2 ENST00000591889.1 |
SPINT2
CTB-102L5.4
|
serine peptidase inhibitor, Kunitz type, 2 CTB-102L5.4 |
| chr4_-_34041504 | 0.31 |
ENST00000512581.1
ENST00000505018.1 |
RP11-79E3.3
|
RP11-79E3.3 |
| chr2_+_143635222 | 0.31 |
ENST00000375773.2
ENST00000409512.1 ENST00000410015.2 |
KYNU
|
kynureninase |
| chr1_-_89591749 | 0.31 |
ENST00000370466.3
|
GBP2
|
guanylate binding protein 2, interferon-inducible |
| chr3_+_52245458 | 0.31 |
ENST00000459884.1
|
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chr12_-_6451235 | 0.31 |
ENST00000440083.2
ENST00000162749.2 |
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A |
| chr11_-_111782696 | 0.31 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr1_-_52870104 | 0.30 |
ENST00000371568.3
|
ORC1
|
origin recognition complex, subunit 1 |
| chr8_+_132952112 | 0.30 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr2_-_166060552 | 0.30 |
ENST00000283254.7
ENST00000453007.1 |
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr5_-_45696253 | 0.30 |
ENST00000303230.4
|
HCN1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr14_-_105487381 | 0.30 |
ENST00000392590.3
ENST00000336219.3 |
CDCA4
|
cell division cycle associated 4 |
| chr11_+_12308447 | 0.30 |
ENST00000256186.2
|
MICALCL
|
MICAL C-terminal like |
| chr4_-_83769996 | 0.29 |
ENST00000511338.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr2_+_161993465 | 0.29 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr17_-_29648761 | 0.29 |
ENST00000247270.3
ENST00000462804.2 |
EVI2A
|
ecotropic viral integration site 2A |
| chr11_-_89653576 | 0.29 |
ENST00000420869.1
|
TRIM49D1
|
tripartite motif containing 49D1 |
| chr13_-_20080080 | 0.29 |
ENST00000400103.2
|
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr21_-_34185989 | 0.29 |
ENST00000487113.1
ENST00000382373.4 |
C21orf62
|
chromosome 21 open reading frame 62 |
| chr2_-_69098566 | 0.29 |
ENST00000295379.1
|
BMP10
|
bone morphogenetic protein 10 |
| chr16_+_56969284 | 0.29 |
ENST00000568358.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr9_-_134585221 | 0.29 |
ENST00000372190.3
ENST00000427994.1 |
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr5_+_162887556 | 0.28 |
ENST00000393915.4
ENST00000432118.2 ENST00000358715.3 |
HMMR
|
hyaluronan-mediated motility receptor (RHAMM) |
| chr2_-_151428735 | 0.28 |
ENST00000441356.1
|
AC104777.2
|
AC104777.2 |
| chr8_+_27631903 | 0.28 |
ENST00000305188.8
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr10_+_127661942 | 0.28 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr4_-_170924888 | 0.28 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr10_+_94352956 | 0.28 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr19_+_38085731 | 0.28 |
ENST00000589117.1
|
ZNF540
|
zinc finger protein 540 |
| chr22_+_24030321 | 0.27 |
ENST00000401461.1
|
RGL4
|
ral guanine nucleotide dissociation stimulator-like 4 |
| chr7_+_44240520 | 0.27 |
ENST00000496112.1
ENST00000223369.2 |
YKT6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr15_+_69706585 | 0.27 |
ENST00000559279.1
ENST00000395392.2 |
KIF23
|
kinesin family member 23 |
| chr15_-_56757329 | 0.27 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr2_+_101591314 | 0.27 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr1_-_53608249 | 0.27 |
ENST00000371494.4
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr12_+_8309630 | 0.27 |
ENST00000396570.3
|
ZNF705A
|
zinc finger protein 705A |
| chr5_+_169010638 | 0.27 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr9_+_100174232 | 0.26 |
ENST00000355295.4
|
TDRD7
|
tudor domain containing 7 |
| chr1_-_9714644 | 0.26 |
ENST00000377320.3
|
C1orf200
|
chromosome 1 open reading frame 200 |
| chr16_+_21689835 | 0.26 |
ENST00000286149.4
ENST00000388958.3 |
OTOA
|
otoancorin |
| chr6_+_22569784 | 0.26 |
ENST00000510882.2
|
HDGFL1
|
hepatoma derived growth factor-like 1 |
| chr5_-_95297678 | 0.26 |
ENST00000237853.4
|
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr6_+_29407083 | 0.26 |
ENST00000444197.2
|
OR10C1
|
olfactory receptor, family 10, subfamily C, member 1 (gene/pseudogene) |
| chr19_+_41281282 | 0.26 |
ENST00000263369.3
|
MIA
|
melanoma inhibitory activity |
| chrX_+_30261847 | 0.26 |
ENST00000378981.3
ENST00000397550.1 |
MAGEB1
|
melanoma antigen family B, 1 |
| chr12_-_21487829 | 0.25 |
ENST00000445053.1
ENST00000452078.1 ENST00000458504.1 ENST00000422327.1 ENST00000421294.1 |
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2 |
| chr18_+_32173276 | 0.25 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr10_-_127371675 | 0.25 |
ENST00000532135.1
ENST00000526819.1 ENST00000368821.3 |
TEX36
|
testis expressed 36 |
| chr3_-_157251383 | 0.25 |
ENST00000487753.1
ENST00000489602.1 ENST00000461299.1 ENST00000479987.1 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr5_+_32788945 | 0.25 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chr15_+_52155001 | 0.25 |
ENST00000544199.1
|
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr8_-_134072593 | 0.25 |
ENST00000427060.2
|
SLA
|
Src-like-adaptor |
| chr6_-_27835357 | 0.25 |
ENST00000331442.3
|
HIST1H1B
|
histone cluster 1, H1b |
| chr7_+_144052381 | 0.25 |
ENST00000498580.1
ENST00000056217.5 |
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5 |
| chr4_-_103997862 | 0.24 |
ENST00000394785.3
|
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr20_-_18477862 | 0.24 |
ENST00000337227.4
|
RBBP9
|
retinoblastoma binding protein 9 |
| chr4_-_47983519 | 0.24 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr5_+_59783540 | 0.24 |
ENST00000515734.2
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr2_+_9983483 | 0.24 |
ENST00000263663.5
|
TAF1B
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63kDa |
| chr3_-_72897545 | 0.24 |
ENST00000325599.8
|
SHQ1
|
SHQ1, H/ACA ribonucleoprotein assembly factor |
| chr5_+_81601166 | 0.24 |
ENST00000439350.1
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr6_+_126277842 | 0.24 |
ENST00000229633.5
|
HINT3
|
histidine triad nucleotide binding protein 3 |
| chr20_+_34824355 | 0.23 |
ENST00000397286.3
ENST00000320849.4 ENST00000373932.3 |
AAR2
|
AAR2 splicing factor homolog (S. cerevisiae) |
| chr8_-_101962777 | 0.23 |
ENST00000395951.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr19_-_48059113 | 0.23 |
ENST00000391901.3
ENST00000314121.4 ENST00000448976.1 |
ZNF541
|
zinc finger protein 541 |
| chr8_+_22225041 | 0.23 |
ENST00000289952.5
ENST00000524285.1 |
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr2_+_113885138 | 0.23 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr2_+_106679690 | 0.23 |
ENST00000409944.1
|
C2orf40
|
chromosome 2 open reading frame 40 |
| chr20_+_57875758 | 0.23 |
ENST00000395654.3
|
EDN3
|
endothelin 3 |
| chr1_-_47407097 | 0.23 |
ENST00000457840.2
|
CYP4A11
|
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr11_-_27384737 | 0.23 |
ENST00000317945.6
|
CCDC34
|
coiled-coil domain containing 34 |
| chr14_+_52164820 | 0.23 |
ENST00000554167.1
|
FRMD6
|
FERM domain containing 6 |
| chr1_+_44584522 | 0.23 |
ENST00000372299.3
|
KLF17
|
Kruppel-like factor 17 |
| chr20_-_1373682 | 0.22 |
ENST00000381724.3
|
FKBP1A
|
FK506 binding protein 1A, 12kDa |
| chr17_+_35732955 | 0.22 |
ENST00000300618.4
|
C17orf78
|
chromosome 17 open reading frame 78 |
| chr14_+_96722152 | 0.22 |
ENST00000216629.6
|
BDKRB1
|
bradykinin receptor B1 |
| chr16_+_1578674 | 0.22 |
ENST00000253934.5
|
TMEM204
|
transmembrane protein 204 |
| chr4_+_169842707 | 0.22 |
ENST00000503290.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr6_-_32784687 | 0.22 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr3_-_46068969 | 0.22 |
ENST00000542109.1
ENST00000395946.2 |
XCR1
|
chemokine (C motif) receptor 1 |
| chr20_+_43990576 | 0.22 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr17_+_34639793 | 0.22 |
ENST00000394465.2
ENST00000394463.2 ENST00000378342.4 |
CCL4L2
|
chemokine (C-C motif) ligand 4-like 2 |
| chr16_+_78056412 | 0.22 |
ENST00000299642.4
ENST00000575655.1 |
CLEC3A
|
C-type lectin domain family 3, member A |
| chr6_+_63921399 | 0.22 |
ENST00000356170.3
|
FKBP1C
|
FK506 binding protein 1C |
| chr5_-_131879205 | 0.22 |
ENST00000231454.1
|
IL5
|
interleukin 5 (colony-stimulating factor, eosinophil) |
| chrX_+_49969405 | 0.22 |
ENST00000376042.1
|
CCNB3
|
cyclin B3 |
| chr12_+_75874460 | 0.21 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr13_-_95248511 | 0.21 |
ENST00000261296.5
|
TGDS
|
TDP-glucose 4,6-dehydratase |
| chr1_-_182369751 | 0.21 |
ENST00000367565.1
|
TEDDM1
|
transmembrane epididymal protein 1 |
| chr6_-_26199471 | 0.21 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr9_+_96846740 | 0.21 |
ENST00000288976.3
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chrX_-_83757399 | 0.21 |
ENST00000373177.2
ENST00000297977.5 ENST00000506585.2 ENST00000449553.2 |
HDX
|
highly divergent homeobox |
| chr10_+_52094298 | 0.21 |
ENST00000595931.1
|
AC069547.1
|
HCG1745369; PRO3073; Uncharacterized protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.0 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 0.8 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.2 | 2.0 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.2 | 0.6 | GO:0032831 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.2 | 0.8 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.2 | 1.4 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 0.5 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.2 | 0.5 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.2 | 0.8 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.4 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 0.8 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.4 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 0.5 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.1 | 1.2 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 1.3 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.5 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.5 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.5 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 0.3 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 0.3 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 1.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.5 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.5 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.6 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.7 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.5 | GO:0048749 | compound eye development(GO:0048749) |
| 0.1 | 0.4 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 1.2 | GO:0014820 | tonic smooth muscle contraction(GO:0014820) |
| 0.1 | 1.0 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 1.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.3 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.1 | 0.3 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 0.6 | GO:0002786 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) negative regulation of defense response to bacterium(GO:1900425) |
| 0.1 | 0.5 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 0.2 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.7 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.2 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.6 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.9 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 1.4 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.1 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 0.3 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.2 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 | 0.2 | GO:0001189 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.1 | 0.2 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.1 | 0.2 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.1 | 0.3 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 1.4 | GO:0060716 | spongiotrophoblast layer development(GO:0060712) labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 0.2 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.3 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.0 | 0.2 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.2 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.1 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.0 | 0.5 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0071048 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.2 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.2 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.2 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.0 | 0.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0002586 | regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002580) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) detection of peptidoglycan(GO:0032499) positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.3 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.3 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.0 | GO:1902567 | negative regulation of eosinophil activation(GO:1902567) |
| 0.0 | 0.2 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.3 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.1 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.5 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.2 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.2 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.2 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.1 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.0 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.3 | GO:0071415 | cellular response to purine-containing compound(GO:0071415) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.6 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 0.2 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.2 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:2000501 | regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.0 | 0.3 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.4 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.8 | GO:2000816 | negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.0 | 0.3 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:0060424 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 0.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.0 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.1 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.2 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.8 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.2 | GO:0015684 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 0.6 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 0.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.1 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 1.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.3 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.3 | GO:0044241 | lipid digestion(GO:0044241) |
| 0.0 | 0.2 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.0 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.0 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.0 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.0 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.0 | 0.0 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 1.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:0098706 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.1 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.0 | 0.1 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.0 | 0.1 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.1 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.2 | 1.4 | GO:1990393 | 3M complex(GO:1990393) |
| 0.2 | 0.5 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.2 | 0.5 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.1 | 0.8 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.3 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.1 | 0.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.5 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 0.3 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.3 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.1 | 0.2 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.1 | 0.5 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.1 | 0.6 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.3 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.7 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.7 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.5 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.1 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.0 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 1.0 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 1.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.2 | 1.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 0.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 1.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 3.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 1.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.2 | 0.8 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 1.0 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.6 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.1 | 0.4 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 0.5 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 0.6 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 0.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.3 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.3 | GO:0038047 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.1 | 0.3 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.1 | 0.2 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 2.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.3 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.1 | 0.2 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 0.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 2.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.4 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.2 | GO:0004040 | amidase activity(GO:0004040) |
| 0.1 | 0.2 | GO:0019961 | interferon binding(GO:0019961) |
| 0.1 | 0.5 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.9 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 2.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.3 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.4 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.2 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.2 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 1.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.5 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.2 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.0 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.7 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.4 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.4 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0030109 | HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.3 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.2 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 1.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.1 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.2 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 1.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.0 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.0 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.0 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.7 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 2.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 3.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 2.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 2.1 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.5 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |