Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
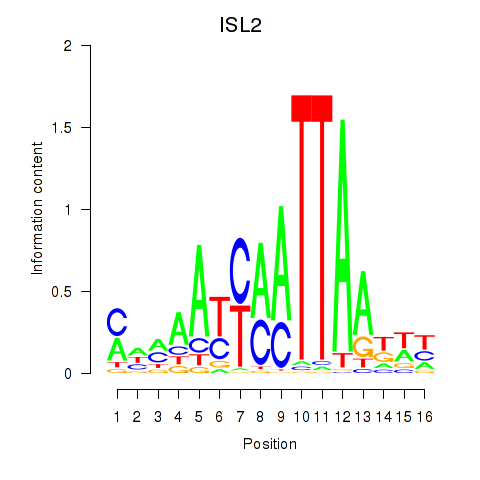
Results for ISL2
Z-value: 0.36
Transcription factors associated with ISL2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ISL2
|
ENSG00000159556.5 | ISL LIM homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ISL2 | hg19_v2_chr15_+_76629064_76629073 | -0.05 | 8.1e-01 | Click! |
Activity profile of ISL2 motif
Sorted Z-values of ISL2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_58212487 | 0.55 |
ENST00000549994.1
|
AVIL
|
advillin |
| chr2_+_86669118 | 0.52 |
ENST00000427678.1
ENST00000542128.1 |
KDM3A
|
lysine (K)-specific demethylase 3A |
| chr4_+_128702969 | 0.50 |
ENST00000508776.1
ENST00000439123.2 |
HSPA4L
|
heat shock 70kDa protein 4-like |
| chr12_+_131438443 | 0.49 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chr7_+_13141097 | 0.47 |
ENST00000411542.1
|
AC011288.2
|
AC011288.2 |
| chr4_+_95128748 | 0.36 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr21_-_31859755 | 0.36 |
ENST00000334055.3
|
KRTAP19-2
|
keratin associated protein 19-2 |
| chr2_+_105050794 | 0.35 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr3_-_141747950 | 0.34 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr7_-_115670804 | 0.33 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr7_-_115670792 | 0.33 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr11_+_5410607 | 0.29 |
ENST00000328611.3
|
OR51M1
|
olfactory receptor, family 51, subfamily M, member 1 |
| chr13_+_78315295 | 0.28 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr14_+_38677123 | 0.27 |
ENST00000267377.2
|
SSTR1
|
somatostatin receptor 1 |
| chr11_-_28129656 | 0.27 |
ENST00000263181.6
|
KIF18A
|
kinesin family member 18A |
| chr1_+_149871135 | 0.27 |
ENST00000369152.5
|
BOLA1
|
bolA family member 1 |
| chr11_+_28129795 | 0.26 |
ENST00000406787.3
ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15
|
methyltransferase like 15 |
| chr17_-_3496171 | 0.25 |
ENST00000399756.4
|
TRPV1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr3_-_191000172 | 0.24 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr14_-_67878917 | 0.23 |
ENST00000216446.4
|
PLEK2
|
pleckstrin 2 |
| chr6_-_111804905 | 0.21 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr11_+_86085778 | 0.18 |
ENST00000354755.1
ENST00000278487.3 ENST00000531271.1 ENST00000445632.2 |
CCDC81
|
coiled-coil domain containing 81 |
| chr12_+_64798095 | 0.17 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chrX_-_32173579 | 0.16 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr4_-_138453606 | 0.16 |
ENST00000412923.2
ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18
|
protocadherin 18 |
| chr2_-_231860596 | 0.15 |
ENST00000441063.1
ENST00000434094.1 ENST00000418330.1 ENST00000457803.1 ENST00000414876.1 ENST00000446741.1 ENST00000426904.1 |
AC105344.2
|
SPATA3 antisense RNA 1 (head to head) |
| chr1_-_190446759 | 0.15 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr5_+_32788945 | 0.14 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chr12_-_95945246 | 0.14 |
ENST00000258499.3
|
USP44
|
ubiquitin specific peptidase 44 |
| chr10_-_128359074 | 0.14 |
ENST00000544758.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr14_+_102276132 | 0.14 |
ENST00000350249.3
ENST00000557621.1 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr4_+_95128996 | 0.14 |
ENST00000457823.2
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr4_+_169013666 | 0.13 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr12_-_118628350 | 0.13 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr14_+_45605157 | 0.13 |
ENST00000542564.2
|
FANCM
|
Fanconi anemia, complementation group M |
| chr6_-_24358264 | 0.13 |
ENST00000378454.3
|
DCDC2
|
doublecortin domain containing 2 |
| chr10_-_5046042 | 0.13 |
ENST00000421196.3
ENST00000455190.1 |
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chr6_+_31105426 | 0.13 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr11_-_124190184 | 0.13 |
ENST00000357438.2
|
OR8D2
|
olfactory receptor, family 8, subfamily D, member 2 |
| chr9_-_99540328 | 0.13 |
ENST00000223428.4
ENST00000375231.1 ENST00000374641.3 |
ZNF510
|
zinc finger protein 510 |
| chr6_+_130339710 | 0.12 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr17_-_10372875 | 0.12 |
ENST00000255381.2
|
MYH4
|
myosin, heavy chain 4, skeletal muscle |
| chr17_+_4710391 | 0.11 |
ENST00000263088.6
ENST00000572940.1 |
PLD2
|
phospholipase D2 |
| chr18_-_64271316 | 0.11 |
ENST00000540086.1
ENST00000580157.1 |
CDH19
|
cadherin 19, type 2 |
| chr2_-_10587897 | 0.11 |
ENST00000405333.1
ENST00000443218.1 |
ODC1
|
ornithine decarboxylase 1 |
| chrX_+_65384182 | 0.10 |
ENST00000441993.2
ENST00000419594.1 |
HEPH
|
hephaestin |
| chr17_-_4938712 | 0.10 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr1_-_232651312 | 0.10 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr8_+_62747349 | 0.10 |
ENST00000517953.1
ENST00000520097.1 ENST00000519766.1 |
RP11-705O24.1
|
RP11-705O24.1 |
| chr6_+_151561085 | 0.10 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr17_-_7307358 | 0.10 |
ENST00000576017.1
ENST00000302422.3 ENST00000535512.1 |
TMEM256
TMEM256-PLSCR3
|
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr19_+_4402659 | 0.09 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chr2_+_234668894 | 0.09 |
ENST00000305208.5
ENST00000608383.1 ENST00000360418.3 |
UGT1A8
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr5_+_68860949 | 0.09 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr1_-_223853348 | 0.09 |
ENST00000366872.5
|
CAPN8
|
calpain 8 |
| chrM_+_9207 | 0.09 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr21_+_39644395 | 0.08 |
ENST00000398934.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr15_-_42500351 | 0.08 |
ENST00000348544.4
ENST00000318006.5 |
VPS39
|
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr1_-_93257951 | 0.08 |
ENST00000543509.1
ENST00000370331.1 ENST00000540033.1 |
EVI5
|
ecotropic viral integration site 5 |
| chr4_+_88532028 | 0.08 |
ENST00000282478.7
|
DSPP
|
dentin sialophosphoprotein |
| chr18_+_61575200 | 0.08 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr5_-_130500922 | 0.08 |
ENST00000513012.1
ENST00000508488.1 ENST00000506908.1 ENST00000304043.5 |
HINT1
|
histidine triad nucleotide binding protein 1 |
| chr5_+_40909354 | 0.08 |
ENST00000313164.9
|
C7
|
complement component 7 |
| chr6_-_155776966 | 0.08 |
ENST00000159060.2
|
NOX3
|
NADPH oxidase 3 |
| chrX_-_15332665 | 0.08 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr2_+_32502952 | 0.08 |
ENST00000238831.4
|
YIPF4
|
Yip1 domain family, member 4 |
| chr16_+_53133070 | 0.08 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr7_-_45151272 | 0.08 |
ENST00000461363.1
ENST00000495078.1 ENST00000494076.1 ENST00000478532.1 ENST00000258770.3 ENST00000361278.3 |
TBRG4
|
transforming growth factor beta regulator 4 |
| chr21_+_39644305 | 0.08 |
ENST00000398930.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chrX_-_77225135 | 0.07 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr1_+_234765057 | 0.07 |
ENST00000429269.1
|
LINC00184
|
long intergenic non-protein coding RNA 184 |
| chr16_-_3350614 | 0.07 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr9_-_21482312 | 0.06 |
ENST00000448696.3
|
IFNE
|
interferon, epsilon |
| chrX_-_119077695 | 0.06 |
ENST00000371410.3
|
NKAP
|
NFKB activating protein |
| chr1_-_12946025 | 0.06 |
ENST00000235349.5
|
PRAMEF4
|
PRAME family member 4 |
| chrX_+_65382433 | 0.06 |
ENST00000374727.3
|
HEPH
|
hephaestin |
| chr1_-_228613026 | 0.06 |
ENST00000366696.1
|
HIST3H3
|
histone cluster 3, H3 |
| chr1_-_13390765 | 0.06 |
ENST00000357367.2
|
PRAMEF8
|
PRAME family member 8 |
| chr10_+_63422695 | 0.06 |
ENST00000330194.2
ENST00000389639.3 |
C10orf107
|
chromosome 10 open reading frame 107 |
| chr4_-_138453559 | 0.06 |
ENST00000511115.1
|
PCDH18
|
protocadherin 18 |
| chr6_+_64345698 | 0.06 |
ENST00000506783.1
ENST00000481385.2 ENST00000515594.1 ENST00000494284.2 ENST00000262043.3 |
PHF3
|
PHD finger protein 3 |
| chr5_+_40841276 | 0.06 |
ENST00000254691.5
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr6_+_29555683 | 0.06 |
ENST00000383640.2
|
OR2H2
|
olfactory receptor, family 2, subfamily H, member 2 |
| chr9_-_100854838 | 0.05 |
ENST00000538344.1
|
TRIM14
|
tripartite motif containing 14 |
| chr14_-_74025625 | 0.05 |
ENST00000553558.1
ENST00000563329.1 ENST00000334988.2 ENST00000560393.1 |
HEATR4
|
HEAT repeat containing 4 |
| chr10_-_95242044 | 0.05 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr18_-_64271363 | 0.05 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr10_-_50396407 | 0.05 |
ENST00000374153.2
ENST00000374151.3 |
C10orf128
|
chromosome 10 open reading frame 128 |
| chr1_+_171227069 | 0.05 |
ENST00000354841.4
|
FMO1
|
flavin containing monooxygenase 1 |
| chr1_+_13421176 | 0.05 |
ENST00000376152.1
|
PRAMEF9
|
PRAME family member 9 |
| chr6_-_152639479 | 0.05 |
ENST00000356820.4
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr3_-_193096600 | 0.05 |
ENST00000446087.1
ENST00000342358.4 |
ATP13A5
|
ATPase type 13A5 |
| chr12_+_26164645 | 0.05 |
ENST00000542004.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr11_+_71544246 | 0.05 |
ENST00000328698.1
|
DEFB108B
|
defensin, beta 108B |
| chr15_-_72563585 | 0.05 |
ENST00000287196.9
ENST00000260376.7 |
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr8_-_121457608 | 0.05 |
ENST00000306185.3
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr3_+_172468472 | 0.04 |
ENST00000232458.5
ENST00000392692.3 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr21_+_15588499 | 0.04 |
ENST00000400577.3
|
RBM11
|
RNA binding motif protein 11 |
| chr17_-_64225508 | 0.04 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr3_-_20053741 | 0.04 |
ENST00000389050.4
|
PP2D1
|
protein phosphatase 2C-like domain containing 1 |
| chr19_-_36001113 | 0.04 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr14_-_53258314 | 0.04 |
ENST00000216410.3
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr4_+_113568207 | 0.04 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr6_+_26440700 | 0.04 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr1_-_168464875 | 0.04 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr2_-_87248975 | 0.04 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr8_-_30706608 | 0.04 |
ENST00000256246.2
|
TEX15
|
testis expressed 15 |
| chr1_+_99127225 | 0.04 |
ENST00000370189.5
ENST00000529992.1 |
SNX7
|
sorting nexin 7 |
| chr13_+_24144509 | 0.03 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr17_-_10325261 | 0.03 |
ENST00000403437.2
|
MYH8
|
myosin, heavy chain 8, skeletal muscle, perinatal |
| chr3_+_172468505 | 0.03 |
ENST00000427830.1
ENST00000417960.1 ENST00000428567.1 ENST00000366090.2 ENST00000426894.1 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr11_-_5323226 | 0.03 |
ENST00000380224.1
|
OR51B4
|
olfactory receptor, family 51, subfamily B, member 4 |
| chrX_+_108779004 | 0.03 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr10_-_95241951 | 0.03 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr2_-_209010874 | 0.03 |
ENST00000260988.4
|
CRYGB
|
crystallin, gamma B |
| chr15_-_83378638 | 0.03 |
ENST00000261722.3
|
AP3B2
|
adaptor-related protein complex 3, beta 2 subunit |
| chr21_-_31864275 | 0.03 |
ENST00000334063.4
|
KRTAP19-3
|
keratin associated protein 19-3 |
| chr3_+_169629354 | 0.03 |
ENST00000428432.2
ENST00000335556.3 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr12_-_54689532 | 0.03 |
ENST00000540264.2
ENST00000312156.4 |
NFE2
|
nuclear factor, erythroid 2 |
| chr21_-_30445886 | 0.03 |
ENST00000431234.1
ENST00000540844.1 ENST00000286788.4 |
CCT8
|
chaperonin containing TCP1, subunit 8 (theta) |
| chr8_-_125577940 | 0.03 |
ENST00000519168.1
ENST00000395508.2 |
MTSS1
|
metastasis suppressor 1 |
| chr4_+_15471489 | 0.03 |
ENST00000424120.1
ENST00000413206.1 ENST00000438599.2 ENST00000511544.1 ENST00000512702.1 ENST00000507954.1 ENST00000515124.1 ENST00000503292.1 ENST00000503658.1 |
CC2D2A
|
coiled-coil and C2 domain containing 2A |
| chr2_-_217560248 | 0.03 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr1_+_63063152 | 0.02 |
ENST00000371129.3
|
ANGPTL3
|
angiopoietin-like 3 |
| chr2_-_217559517 | 0.02 |
ENST00000449583.1
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr19_-_51920952 | 0.02 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr12_-_10282836 | 0.02 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chrX_+_102840408 | 0.02 |
ENST00000468024.1
ENST00000472484.1 ENST00000415568.2 ENST00000490644.1 ENST00000459722.1 ENST00000472745.1 ENST00000494801.1 ENST00000434216.2 ENST00000425011.1 |
TCEAL4
|
transcription elongation factor A (SII)-like 4 |
| chr10_-_27529486 | 0.02 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr2_+_113825547 | 0.02 |
ENST00000341010.2
ENST00000337569.3 |
IL1F10
|
interleukin 1 family, member 10 (theta) |
| chr12_+_122356488 | 0.02 |
ENST00000397454.2
|
WDR66
|
WD repeat domain 66 |
| chr4_-_69111401 | 0.02 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr1_+_149871171 | 0.02 |
ENST00000369150.1
|
BOLA1
|
bolA family member 1 |
| chr8_+_92261516 | 0.02 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr13_+_49551020 | 0.02 |
ENST00000541916.1
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr15_+_79166065 | 0.02 |
ENST00000559690.1
ENST00000559158.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr6_+_161123270 | 0.02 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr11_+_12132117 | 0.01 |
ENST00000256194.4
|
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr19_+_29456034 | 0.01 |
ENST00000589921.1
|
LINC00906
|
long intergenic non-protein coding RNA 906 |
| chr11_-_92931098 | 0.01 |
ENST00000326402.4
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chr14_-_90798418 | 0.01 |
ENST00000354366.3
|
NRDE2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr12_-_10282681 | 0.01 |
ENST00000533022.1
|
CLEC7A
|
C-type lectin domain family 7, member A |
| chr2_+_234826016 | 0.01 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr7_+_141811539 | 0.01 |
ENST00000550469.2
ENST00000477922.3 |
RP11-1220K2.2
|
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr11_-_5255861 | 0.01 |
ENST00000380299.3
|
HBD
|
hemoglobin, delta |
| chr18_-_3845321 | 0.01 |
ENST00000539435.1
ENST00000400147.2 |
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr2_-_159237472 | 0.01 |
ENST00000409187.1
|
CCDC148
|
coiled-coil domain containing 148 |
| chr12_-_10282742 | 0.01 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr13_-_45768841 | 0.01 |
ENST00000379108.1
|
KCTD4
|
potassium channel tetramerization domain containing 4 |
| chr5_+_140201183 | 0.01 |
ENST00000529619.1
ENST00000529859.1 ENST00000378126.3 |
PCDHA5
|
protocadherin alpha 5 |
| chr7_-_16844611 | 0.01 |
ENST00000401412.1
ENST00000419304.2 |
AGR2
|
anterior gradient 2 |
| chr1_-_178840157 | 0.00 |
ENST00000367629.1
ENST00000234816.2 |
ANGPTL1
|
angiopoietin-like 1 |
| chr3_-_20227619 | 0.00 |
ENST00000425061.1
ENST00000443724.1 ENST00000421451.1 ENST00000452020.1 ENST00000417364.1 ENST00000306698.2 ENST00000419233.2 ENST00000263753.4 ENST00000383774.1 ENST00000437051.1 ENST00000412868.1 ENST00000429446.3 ENST00000442720.1 |
SGOL1
|
shugoshin-like 1 (S. pombe) |
| chr3_-_157221380 | 0.00 |
ENST00000468233.1
|
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr2_+_88047606 | 0.00 |
ENST00000359481.4
|
PLGLB2
|
plasminogen-like B2 |
| chr14_+_22670455 | 0.00 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr2_+_74685413 | 0.00 |
ENST00000233615.2
|
WBP1
|
WW domain binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ISL2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 0.2 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.1 | 0.3 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.5 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.3 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.0 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.5 | GO:0035861 | site of double-strand break(GO:0035861) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.3 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.2 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |