Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
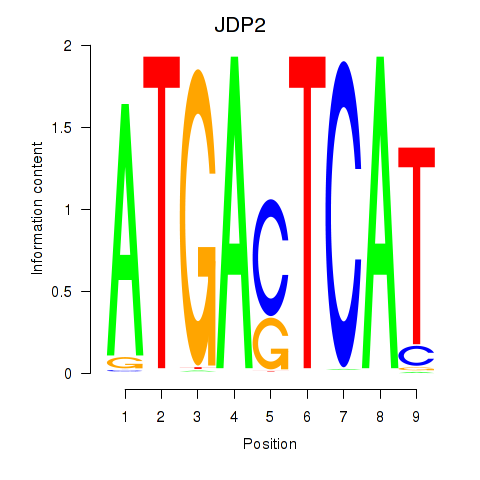
Results for JDP2
Z-value: 0.91
Transcription factors associated with JDP2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JDP2
|
ENSG00000140044.8 | Jun dimerization protein 2 |
Activity profile of JDP2 motif
Sorted Z-values of JDP2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_89182178 | 13.12 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr15_+_89182156 | 12.93 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr15_+_89181974 | 11.38 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr2_+_152214098 | 6.64 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr21_+_26934165 | 6.45 |
ENST00000456917.1
|
MIR155HG
|
MIR155 host gene (non-protein coding) |
| chr4_+_74606223 | 6.01 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr1_-_153521597 | 5.57 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr11_-_102668879 | 5.50 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr1_-_153521714 | 5.04 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr7_+_55177416 | 4.76 |
ENST00000450046.1
ENST00000454757.2 |
EGFR
|
epidermal growth factor receptor |
| chr16_+_57662138 | 3.42 |
ENST00000562414.1
ENST00000561969.1 ENST00000562631.1 ENST00000563445.1 ENST00000565338.1 ENST00000567702.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr16_+_57662419 | 3.40 |
ENST00000388812.4
ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56
|
G protein-coupled receptor 56 |
| chr1_+_183155373 | 3.11 |
ENST00000493293.1
ENST00000264144.4 |
LAMC2
|
laminin, gamma 2 |
| chr8_+_86157699 | 3.11 |
ENST00000321764.3
|
CA13
|
carbonic anhydrase XIII |
| chr5_+_35856951 | 3.06 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr10_+_104155450 | 2.61 |
ENST00000471698.1
ENST00000189444.6 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr4_-_122085469 | 2.30 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr2_-_175712270 | 2.29 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr17_+_74381343 | 2.04 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr11_-_2950642 | 1.97 |
ENST00000314222.4
|
PHLDA2
|
pleckstrin homology-like domain, family A, member 2 |
| chr11_+_122526383 | 1.82 |
ENST00000284273.5
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B |
| chr18_+_21693306 | 1.80 |
ENST00000540918.2
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr18_+_55888767 | 1.78 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr17_-_28257080 | 1.77 |
ENST00000579954.1
ENST00000540801.1 ENST00000269033.3 ENST00000590153.1 ENST00000582084.1 |
SSH2
|
slingshot protein phosphatase 2 |
| chr1_-_95007193 | 1.76 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr7_+_28725585 | 1.69 |
ENST00000396298.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr1_-_209824643 | 1.54 |
ENST00000391911.1
ENST00000415782.1 |
LAMB3
|
laminin, beta 3 |
| chr4_+_86525299 | 1.31 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr6_+_151042224 | 1.21 |
ENST00000358517.2
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr1_+_213224572 | 1.20 |
ENST00000543470.1
ENST00000366960.3 ENST00000366959.3 ENST00000543354.1 |
RPS6KC1
|
ribosomal protein S6 kinase, 52kDa, polypeptide 1 |
| chr11_-_133715394 | 1.17 |
ENST00000299140.3
ENST00000532889.1 |
SPATA19
|
spermatogenesis associated 19 |
| chr21_-_36421535 | 1.13 |
ENST00000416754.1
ENST00000437180.1 ENST00000455571.1 |
RUNX1
|
runt-related transcription factor 1 |
| chr21_-_36421626 | 1.08 |
ENST00000300305.3
|
RUNX1
|
runt-related transcription factor 1 |
| chr3_-_195997410 | 1.06 |
ENST00000419333.1
|
PCYT1A
|
phosphate cytidylyltransferase 1, choline, alpha |
| chr7_+_134528635 | 1.05 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr22_+_39916558 | 1.04 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr1_+_156084461 | 0.99 |
ENST00000347559.2
ENST00000361308.4 ENST00000368300.4 ENST00000368299.3 |
LMNA
|
lamin A/C |
| chr17_+_79650962 | 0.97 |
ENST00000329138.4
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr5_+_179247759 | 0.89 |
ENST00000389805.4
ENST00000504627.1 ENST00000402874.3 ENST00000510187.1 |
SQSTM1
|
sequestosome 1 |
| chr3_+_48507621 | 0.88 |
ENST00000456089.1
|
TREX1
|
three prime repair exonuclease 1 |
| chr6_+_106534192 | 0.86 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr9_+_138453595 | 0.85 |
ENST00000479141.1
ENST00000371766.2 ENST00000277508.5 ENST00000433563.1 |
PAEP
|
progestagen-associated endometrial protein |
| chr6_-_84140757 | 0.85 |
ENST00000541327.1
ENST00000369705.3 ENST00000543031.1 |
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr22_+_20861858 | 0.85 |
ENST00000414658.1
ENST00000432052.1 ENST00000425759.2 ENST00000292733.7 ENST00000542773.1 ENST00000263205.7 ENST00000406969.1 ENST00000382974.2 |
MED15
|
mediator complex subunit 15 |
| chr1_-_111148241 | 0.82 |
ENST00000440270.1
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr14_+_103800513 | 0.80 |
ENST00000560338.1
ENST00000560763.1 ENST00000558316.1 ENST00000558265.1 |
EIF5
|
eukaryotic translation initiation factor 5 |
| chr3_-_191000172 | 0.79 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr9_-_123638633 | 0.79 |
ENST00000456291.1
|
PHF19
|
PHD finger protein 19 |
| chr2_+_69201705 | 0.77 |
ENST00000377938.2
|
GKN1
|
gastrokine 1 |
| chr2_+_74757050 | 0.77 |
ENST00000352222.3
ENST00000437202.1 |
HTRA2
|
HtrA serine peptidase 2 |
| chr12_+_75760714 | 0.77 |
ENST00000547144.1
|
GLIPR1L1
|
GLI pathogenesis-related 1 like 1 |
| chr10_-_120514720 | 0.77 |
ENST00000369151.3
ENST00000340214.4 |
CACUL1
|
CDK2-associated, cullin domain 1 |
| chr3_+_48507210 | 0.77 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr1_+_152881014 | 0.77 |
ENST00000368764.3
ENST00000392667.2 |
IVL
|
involucrin |
| chr9_-_123639304 | 0.75 |
ENST00000436309.1
|
PHF19
|
PHD finger protein 19 |
| chr6_+_45296048 | 0.75 |
ENST00000465038.2
ENST00000352853.5 ENST00000541979.1 ENST00000371438.1 |
RUNX2
|
runt-related transcription factor 2 |
| chrX_-_48901012 | 0.67 |
ENST00000315869.7
|
TFE3
|
transcription factor binding to IGHM enhancer 3 |
| chr17_-_47287928 | 0.67 |
ENST00000507680.1
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr7_+_48128194 | 0.66 |
ENST00000416681.1
ENST00000331803.4 ENST00000432131.1 |
UPP1
|
uridine phosphorylase 1 |
| chr18_-_74839891 | 0.64 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr7_+_48128316 | 0.63 |
ENST00000341253.4
|
UPP1
|
uridine phosphorylase 1 |
| chr4_+_169013666 | 0.63 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr11_+_5009424 | 0.59 |
ENST00000300762.1
|
MMP26
|
matrix metallopeptidase 26 |
| chr1_-_203198790 | 0.58 |
ENST00000367229.1
ENST00000255427.3 ENST00000535569.1 |
CHIT1
|
chitinase 1 (chitotriosidase) |
| chr16_+_30211181 | 0.56 |
ENST00000395138.2
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr17_-_38911580 | 0.56 |
ENST00000312150.4
|
KRT25
|
keratin 25 |
| chr17_-_39296739 | 0.52 |
ENST00000345847.4
|
KRTAP4-6
|
keratin associated protein 4-6 |
| chr17_-_33760269 | 0.51 |
ENST00000452764.3
|
SLFN12
|
schlafen family member 12 |
| chr1_+_156096336 | 0.50 |
ENST00000504687.1
ENST00000473598.2 |
LMNA
|
lamin A/C |
| chr17_+_28256874 | 0.49 |
ENST00000541045.1
ENST00000536908.2 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chrX_+_41193407 | 0.48 |
ENST00000457138.2
ENST00000441189.2 |
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr5_-_149492904 | 0.48 |
ENST00000286301.3
ENST00000511344.1 |
CSF1R
|
colony stimulating factor 1 receptor |
| chr17_+_30771279 | 0.45 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr15_+_22892663 | 0.43 |
ENST00000313077.7
ENST00000561274.1 ENST00000560848.1 |
CYFIP1
|
cytoplasmic FMR1 interacting protein 1 |
| chr16_-_31161380 | 0.43 |
ENST00000569305.1
ENST00000418068.2 ENST00000268281.4 |
PRSS36
|
protease, serine, 36 |
| chr10_-_4285923 | 0.43 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr20_+_30946106 | 0.42 |
ENST00000375687.4
ENST00000542461.1 |
ASXL1
|
additional sex combs like 1 (Drosophila) |
| chr3_+_183903811 | 0.42 |
ENST00000429586.2
ENST00000292808.5 |
ABCF3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr12_-_71182695 | 0.42 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr17_+_4853442 | 0.41 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr5_+_72143988 | 0.41 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr2_-_180610767 | 0.40 |
ENST00000409343.1
|
ZNF385B
|
zinc finger protein 385B |
| chr12_-_131200810 | 0.39 |
ENST00000536002.1
ENST00000544034.1 |
RIMBP2
RP11-662M24.2
|
RIMS binding protein 2 RP11-662M24.2 |
| chrX_+_153770421 | 0.39 |
ENST00000369609.5
ENST00000369607.1 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr6_+_10521574 | 0.38 |
ENST00000495262.1
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr7_-_155604967 | 0.38 |
ENST00000297261.2
|
SHH
|
sonic hedgehog |
| chr1_-_67142710 | 0.36 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr2_-_220173685 | 0.35 |
ENST00000423636.2
ENST00000442029.1 ENST00000412847.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr10_+_88854926 | 0.32 |
ENST00000298784.1
ENST00000298786.4 |
FAM35A
|
family with sequence similarity 35, member A |
| chr11_-_57417405 | 0.32 |
ENST00000524669.1
ENST00000300022.3 |
YPEL4
|
yippee-like 4 (Drosophila) |
| chr2_+_166326157 | 0.31 |
ENST00000421875.1
ENST00000314499.7 ENST00000409664.1 |
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr10_-_31288398 | 0.30 |
ENST00000538351.2
|
ZNF438
|
zinc finger protein 438 |
| chr2_+_12246664 | 0.30 |
ENST00000449986.1
|
AC096559.1
|
AC096559.1 |
| chrY_+_15016725 | 0.29 |
ENST00000336079.3
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr5_+_145317356 | 0.29 |
ENST00000511217.1
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr17_-_1553346 | 0.28 |
ENST00000301336.6
|
RILP
|
Rab interacting lysosomal protein |
| chr2_-_145275228 | 0.28 |
ENST00000427902.1
ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr3_+_98699880 | 0.28 |
ENST00000473756.1
|
LINC00973
|
long intergenic non-protein coding RNA 973 |
| chr12_-_76817036 | 0.28 |
ENST00000546946.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr3_-_48632593 | 0.28 |
ENST00000454817.1
ENST00000328333.8 |
COL7A1
|
collagen, type VII, alpha 1 |
| chr4_+_175839506 | 0.26 |
ENST00000505141.1
ENST00000359240.3 ENST00000445694.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr3_-_49395705 | 0.25 |
ENST00000419349.1
|
GPX1
|
glutathione peroxidase 1 |
| chr1_-_160231451 | 0.25 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr9_+_140135665 | 0.25 |
ENST00000340384.4
|
TUBB4B
|
tubulin, beta 4B class IVb |
| chr7_+_147830776 | 0.25 |
ENST00000538075.1
|
CNTNAP2
|
contactin associated protein-like 2 |
| chr16_+_89988259 | 0.25 |
ENST00000554444.1
ENST00000556565.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr19_-_35992780 | 0.25 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr5_+_177540444 | 0.25 |
ENST00000274605.5
|
N4BP3
|
NEDD4 binding protein 3 |
| chrX_+_47053208 | 0.24 |
ENST00000442035.1
ENST00000457753.1 ENST00000335972.6 |
UBA1
|
ubiquitin-like modifier activating enzyme 1 |
| chr9_-_21335240 | 0.24 |
ENST00000537938.1
|
KLHL9
|
kelch-like family member 9 |
| chr7_-_122342988 | 0.23 |
ENST00000434824.1
|
RNF148
|
ring finger protein 148 |
| chr4_+_175839551 | 0.23 |
ENST00000404450.4
ENST00000514159.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr17_-_47785504 | 0.22 |
ENST00000514907.1
ENST00000503334.1 ENST00000508520.1 |
SLC35B1
|
solute carrier family 35, member B1 |
| chr1_-_166944561 | 0.22 |
ENST00000271417.3
|
ILDR2
|
immunoglobulin-like domain containing receptor 2 |
| chr9_-_23826298 | 0.22 |
ENST00000380117.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr19_-_46285646 | 0.22 |
ENST00000458663.2
|
DMPK
|
dystrophia myotonica-protein kinase |
| chr13_-_41593425 | 0.21 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chrX_-_153285395 | 0.21 |
ENST00000369980.3
|
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr16_-_69760409 | 0.21 |
ENST00000561500.1
ENST00000439109.2 ENST00000564043.1 ENST00000379046.2 ENST00000379047.3 |
NQO1
|
NAD(P)H dehydrogenase, quinone 1 |
| chr8_-_82395461 | 0.20 |
ENST00000256104.4
|
FABP4
|
fatty acid binding protein 4, adipocyte |
| chrX_-_153285251 | 0.20 |
ENST00000444230.1
ENST00000393682.1 ENST00000393687.2 ENST00000429936.2 ENST00000369974.2 |
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr14_+_103801140 | 0.20 |
ENST00000561325.1
ENST00000392715.2 ENST00000559130.1 ENST00000559532.1 ENST00000558506.1 |
EIF5
|
eukaryotic translation initiation factor 5 |
| chr9_+_44867571 | 0.20 |
ENST00000377548.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr7_-_122342966 | 0.20 |
ENST00000447240.1
|
RNF148
|
ring finger protein 148 |
| chr12_-_6960407 | 0.19 |
ENST00000540683.1
ENST00000229265.6 ENST00000535406.1 ENST00000422785.3 |
CDCA3
|
cell division cycle associated 3 |
| chr2_-_74757066 | 0.18 |
ENST00000377526.3
|
AUP1
|
ancient ubiquitous protein 1 |
| chr14_+_73525144 | 0.18 |
ENST00000261973.7
ENST00000540173.1 |
RBM25
|
RNA binding motif protein 25 |
| chr10_+_121410882 | 0.17 |
ENST00000369085.3
|
BAG3
|
BCL2-associated athanogene 3 |
| chrX_+_57618269 | 0.17 |
ENST00000374888.1
|
ZXDB
|
zinc finger, X-linked, duplicated B |
| chr9_-_91793675 | 0.17 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr3_-_45957088 | 0.17 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr3_-_45957534 | 0.16 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr14_+_73525229 | 0.16 |
ENST00000527432.1
ENST00000531500.1 ENST00000525321.1 ENST00000526754.1 |
RBM25
|
RNA binding motif protein 25 |
| chr1_+_151227179 | 0.16 |
ENST00000368884.3
ENST00000368881.4 |
PSMD4
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 |
| chr7_-_14942283 | 0.15 |
ENST00000402815.1
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chrX_-_70326455 | 0.14 |
ENST00000374251.5
|
CXorf65
|
chromosome X open reading frame 65 |
| chr4_-_36246060 | 0.14 |
ENST00000303965.4
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr6_-_49755019 | 0.14 |
ENST00000304801.3
|
PGK2
|
phosphoglycerate kinase 2 |
| chr3_+_191046810 | 0.14 |
ENST00000392455.3
ENST00000392456.3 |
CCDC50
|
coiled-coil domain containing 50 |
| chr3_+_155860751 | 0.14 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr14_-_51561784 | 0.14 |
ENST00000360392.4
|
TRIM9
|
tripartite motif containing 9 |
| chrX_+_133371077 | 0.14 |
ENST00000517294.1
ENST00000370809.4 |
CCDC160
|
coiled-coil domain containing 160 |
| chr14_+_73525265 | 0.13 |
ENST00000525161.1
|
RBM25
|
RNA binding motif protein 25 |
| chr2_+_169926047 | 0.13 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr16_+_3692931 | 0.13 |
ENST00000407479.1
|
DNASE1
|
deoxyribonuclease I |
| chr2_-_11606275 | 0.12 |
ENST00000381525.3
ENST00000362009.4 |
E2F6
|
E2F transcription factor 6 |
| chr11_-_66103867 | 0.12 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr3_+_19988885 | 0.12 |
ENST00000422242.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr1_-_244006528 | 0.11 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr7_-_150721570 | 0.11 |
ENST00000377974.2
ENST00000444312.1 ENST00000605938.1 ENST00000605952.1 |
ATG9B
|
autophagy related 9B |
| chr18_+_2846972 | 0.11 |
ENST00000254528.3
|
EMILIN2
|
elastin microfibril interfacer 2 |
| chr6_+_125540951 | 0.11 |
ENST00000524679.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr1_+_15943995 | 0.11 |
ENST00000480945.1
|
DDI2
|
DNA-damage inducible 1 homolog 2 (S. cerevisiae) |
| chr3_+_138153451 | 0.11 |
ENST00000389567.4
ENST00000289135.4 |
ESYT3
|
extended synaptotagmin-like protein 3 |
| chr6_-_31704282 | 0.10 |
ENST00000375784.3
ENST00000375779.2 |
CLIC1
|
chloride intracellular channel 1 |
| chr15_-_54025300 | 0.10 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr19_+_13842559 | 0.09 |
ENST00000586600.1
|
CCDC130
|
coiled-coil domain containing 130 |
| chr2_+_47596287 | 0.09 |
ENST00000263735.4
|
EPCAM
|
epithelial cell adhesion molecule |
| chr1_+_26606608 | 0.09 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr1_-_13007420 | 0.08 |
ENST00000376189.1
|
PRAMEF6
|
PRAME family member 6 |
| chr19_+_3366547 | 0.08 |
ENST00000341919.3
ENST00000590282.1 ENST00000443272.2 |
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor) |
| chr7_+_23286182 | 0.07 |
ENST00000258733.4
ENST00000381990.2 ENST00000409458.3 ENST00000539136.1 ENST00000453162.2 |
GPNMB
|
glycoprotein (transmembrane) nmb |
| chr16_+_14280564 | 0.07 |
ENST00000572567.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr9_-_21335356 | 0.07 |
ENST00000359039.4
|
KLHL9
|
kelch-like family member 9 |
| chr10_-_90611566 | 0.07 |
ENST00000371930.4
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr11_-_66103932 | 0.07 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr17_-_73150629 | 0.07 |
ENST00000356033.4
ENST00000405458.3 ENST00000409753.3 |
HN1
|
hematological and neurological expressed 1 |
| chrX_-_154493791 | 0.06 |
ENST00000369454.3
|
RAB39B
|
RAB39B, member RAS oncogene family |
| chr1_-_13117736 | 0.06 |
ENST00000376192.5
ENST00000376182.1 |
PRAMEF6
|
PRAME family member 6 |
| chr19_-_51872233 | 0.05 |
ENST00000601435.1
ENST00000291715.1 |
CLDND2
|
claudin domain containing 2 |
| chr12_-_57914275 | 0.04 |
ENST00000547303.1
ENST00000552740.1 ENST00000547526.1 ENST00000551116.1 ENST00000346473.3 |
DDIT3
|
DNA-damage-inducible transcript 3 |
| chr20_-_62129163 | 0.04 |
ENST00000298049.7
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr7_-_55606346 | 0.04 |
ENST00000545390.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr6_+_56819895 | 0.04 |
ENST00000370748.3
|
BEND6
|
BEN domain containing 6 |
| chr12_+_113860160 | 0.04 |
ENST00000553248.1
ENST00000345635.4 ENST00000547802.1 |
SDSL
|
serine dehydratase-like |
| chr12_+_113860042 | 0.03 |
ENST00000403593.4
|
SDSL
|
serine dehydratase-like |
| chr10_+_1102721 | 0.03 |
ENST00000263150.4
|
WDR37
|
WD repeat domain 37 |
| chr2_+_68872954 | 0.03 |
ENST00000394342.2
|
PROKR1
|
prokineticin receptor 1 |
| chr19_-_51538118 | 0.02 |
ENST00000529888.1
|
KLK12
|
kallikrein-related peptidase 12 |
| chr9_+_34958254 | 0.02 |
ENST00000242315.3
|
KIAA1045
|
KIAA1045 |
| chr16_+_30077055 | 0.01 |
ENST00000564595.2
ENST00000569798.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr2_+_233897382 | 0.01 |
ENST00000233840.3
|
NEU2
|
sialidase 2 (cytosolic sialidase) |
| chr18_+_61442629 | 0.01 |
ENST00000398019.2
ENST00000540675.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr4_+_120056939 | 0.01 |
ENST00000307128.5
|
MYOZ2
|
myozenin 2 |
| chr1_+_13359819 | 0.01 |
ENST00000376168.1
|
PRAMEF5
|
PRAME family member 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of JDP2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 37.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.2 | 4.8 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.7 | 2.0 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.7 | 2.0 | GO:0060723 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.7 | 2.6 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.6 | 1.8 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.4 | 1.8 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.3 | 1.0 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.3 | 1.3 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.3 | 1.5 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.3 | 0.9 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 0.3 | 0.8 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.3 | 3.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.3 | 0.8 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.2 | 6.6 | GO:0030728 | ovulation(GO:0030728) |
| 0.2 | 0.9 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.2 | 0.8 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.2 | 4.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.4 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.1 | 0.4 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 2.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 1.8 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 1.5 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.4 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of immature T cell proliferation in thymus(GO:0033092) right lung development(GO:0060458) primary prostatic bud elongation(GO:0060516) epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.1 | 0.2 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 0.8 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.1 | 0.3 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.6 | GO:1904207 | maintenance of blood-brain barrier(GO:0035633) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 4.0 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 0.3 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 1.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.4 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.1 | 0.6 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 0.6 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 1.0 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.7 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.2 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.1 | 0.4 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.5 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.1 | 0.3 | GO:1902903 | regulation of fibril organization(GO:1902903) |
| 0.1 | 3.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 2.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.5 | GO:0090197 | positive regulation of chemokine secretion(GO:0090197) |
| 0.1 | 0.9 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 1.0 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.0 | 0.4 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 2.4 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 1.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.0 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.6 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.2 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 1.0 | 3.1 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.7 | 2.6 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.5 | 37.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.3 | 1.0 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.3 | 0.9 | GO:0044753 | amphisome(GO:0044753) |
| 0.2 | 1.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.2 | 1.0 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.2 | 1.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 1.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.3 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.5 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 6.5 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 1.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.3 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.4 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 1.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 0.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.8 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 1.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 2.9 | GO:0043209 | myelin sheath(GO:0043209) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.5 | 37.4 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.0 | 3.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 1.0 | 4.8 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.3 | 2.0 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.3 | 1.6 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.3 | 1.3 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.2 | 1.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 6.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.0 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.2 | 3.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.8 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 1.8 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.6 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.1 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.2 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) |
| 0.1 | 3.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 1.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.4 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.8 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.5 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.4 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.6 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 4.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.0 | 3.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 1.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 3.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.7 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 3.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 4.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 6.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.0 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 3.1 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 1.7 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 4.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 2.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 9.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 6.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.5 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 37.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 7.1 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.2 | 5.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 3.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 4.8 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.2 | 3.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 3.0 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 0.2 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 1.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 2.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.8 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 4.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 2.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 1.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |